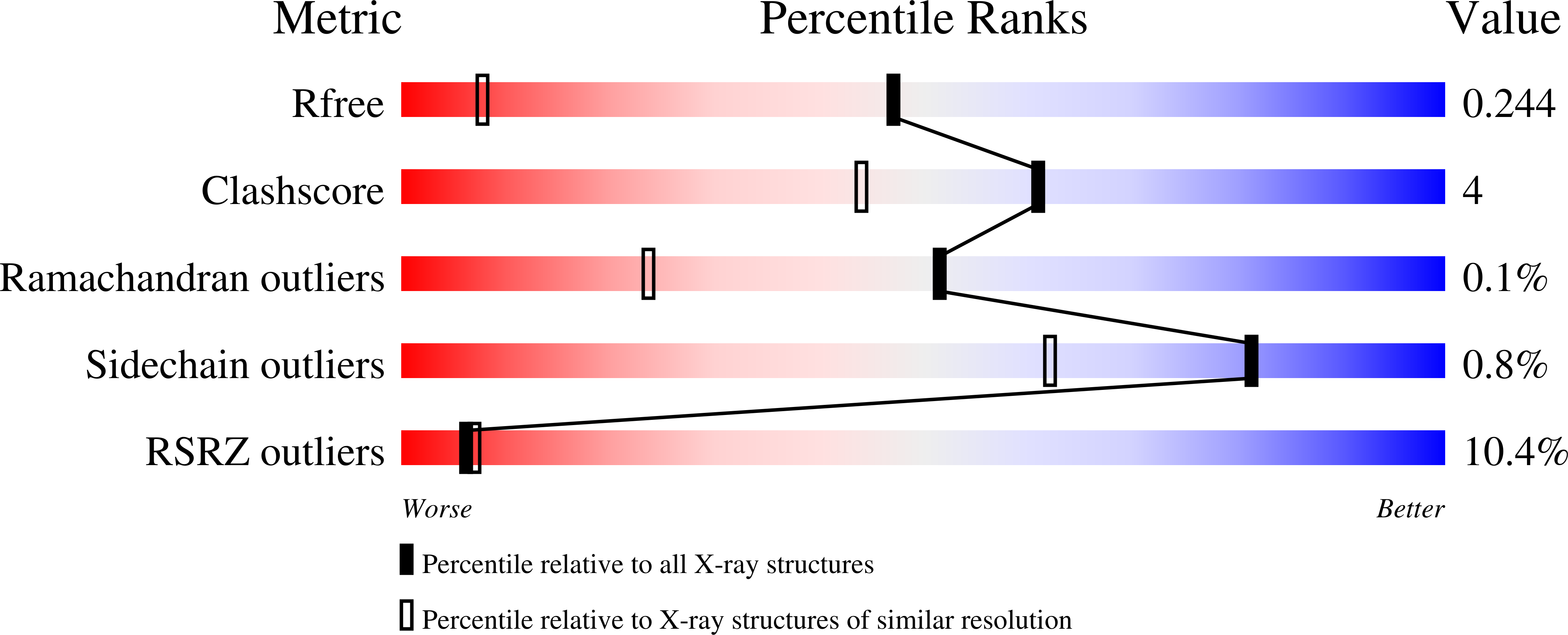
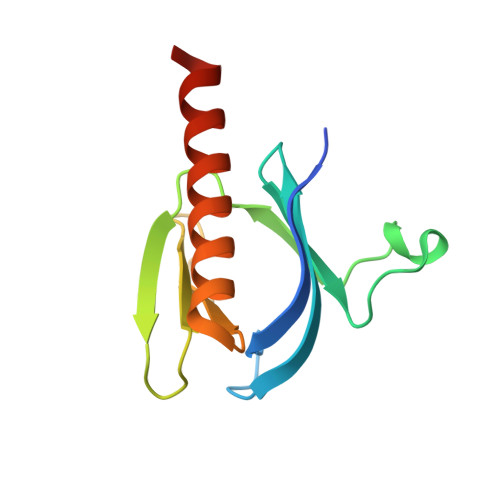
The Crystal Structure of the Ph Domain of Human Akt3 Protein Kinase
Vollmar, M., Wang, J., Zhang, Y., Elkins, J.M., Burgess-Brown, N., Chaikuad, A., Pike, A.C.W., von Delft, F., Bountra, C., Arrowsmith, C.H., Weigelt, J., Edwards, A., Knapp, S.To be published.