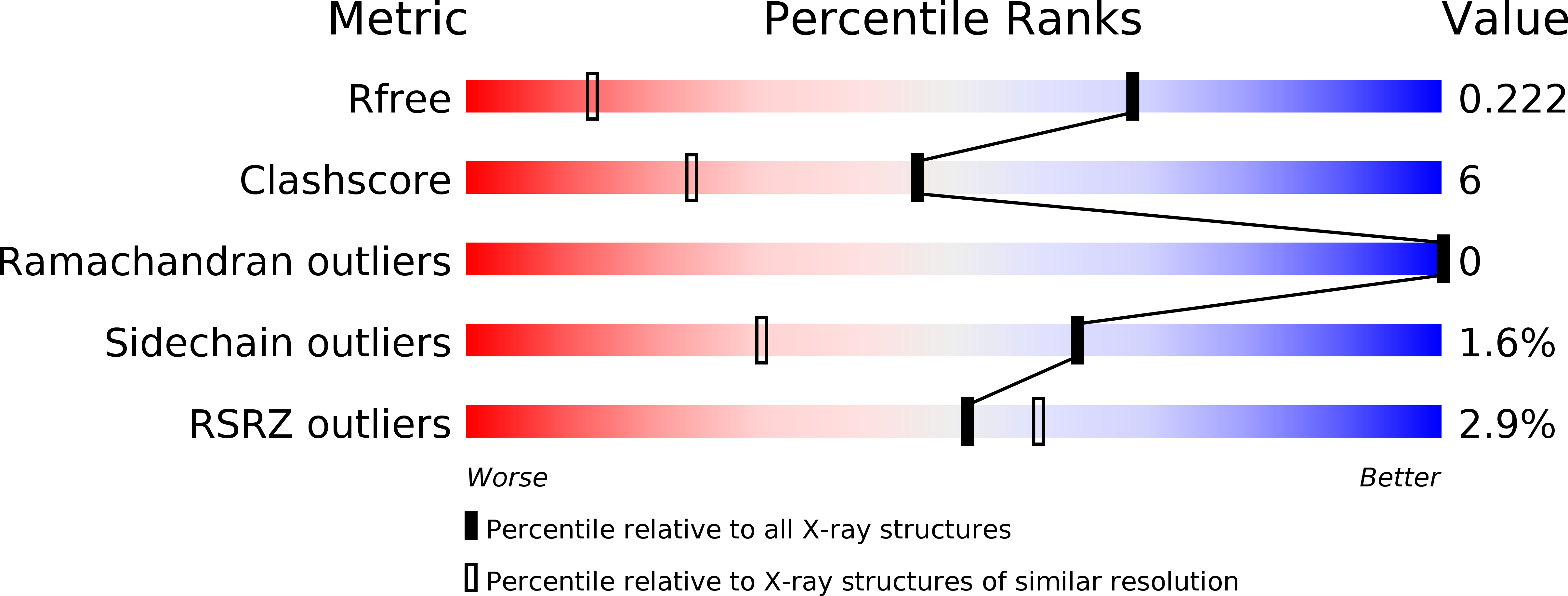
The Single-Domain Globin from the Pathogenic Bacterium Campylobacter Jejuni: Novel D-Helix Conformation, Proximal Hydrogen Bonding that Influences Ligand Binding, and Peroxidase-Like Redox Properties.
Shepherd, M., Barynin, V.V., Lu, C., Bernhardt, P.V., Wu, G., Yeh, S.R., Egawa, T., Sedelnikova, S.E., Rice, D.W., Wilson, J.L., Poole, R.K.(2010) J Biol Chem 285: 12747
- PubMed: 20164176
- DOI: https://doi.org/10.1074/jbc.M109.084509
- Primary Citation of Related Structures:
2WY4 - PubMed Abstract:
The food-borne pathogen Campylobacter jejuni possesses a single-domain globin (Cgb) whose role in detoxifying nitric oxide has been unequivocally demonstrated through genetic and molecular approaches. The x-ray structure of cyanide-bound Cgb has been solved to a resolution of 1.35 A. The overall fold is a classic three-on-three alpha-helical globin fold, similar to that of myoglobin and Vgb from Vitreoscilla stercoraria. However, the D region (defined according to the standard globin fold nomenclature) of Cgb adopts a highly ordered alpha-helical conformation unlike any previously characterized members of this globin family, and the GlnE7 residue has an unexpected role in modulating the interaction between the ligand and the TyrB10 residue. The proximal hydrogen bonding network in Cgb demonstrates that the heme cofactor is ligated by an imidazolate, a characteristic of peroxidase-like proteins. Mutation of either proximal hydrogen-bonding residue (GluH23 or TyrG5) results in the loss of the high frequency nu(Fe-His) stretching mode (251 cm(-1)), indicating that both residues are important for maintaining the anionic character of the proximal histidine ligand. Cyanide binding kinetics for these proximal mutants demonstrate for the first time that proximal hydrogen bonding in globins can modulate ligand binding kinetics at the distal site. A low redox midpoint for the ferrous/ferric couple (-134 mV versus normal hydrogen electrode at pH 7) is consistent with the peroxidase-like character of the Cgb active site. These data provide a new insight into the mechanism via which Campylobacter may survive host-derived nitrosative stress.
Organizational Affiliation:
Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield S10 2TN, United Kingdom. m.shepherd@sheffield.ac.uk