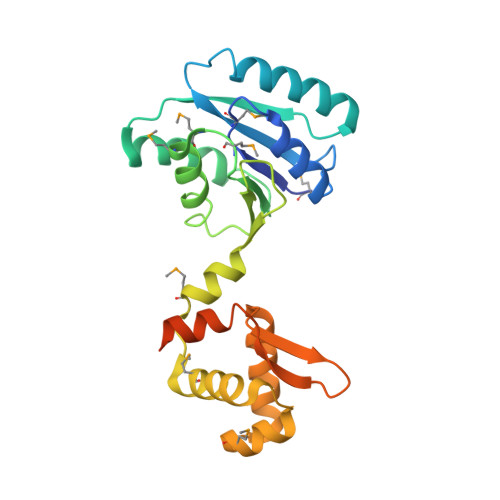
The Structure of the Crispr-Associated Protein Csa3 Provides Insight Into the Regulation of the Crispr/Cas System.
Lintner, N.G., Frankel, K.A., Tsutakawa, S.E., Alsbury, D.L., Copie, V., Young, M.J., Tainer, J.A., Lawrence, C.M.(2011) J Mol Biol 405: 939
- PubMed: 21093452
- DOI: https://doi.org/10.1016/j.jmb.2010.11.019
- Primary Citation of Related Structures:
2WTE - PubMed Abstract:
Adaptive immune systems have recently been recognized in prokaryotic organisms where, in response to viral infection, they incorporate short fragments of invader-derived DNA into loci called clustered regularly interspaced short palindromic repeats (CRISPRs). In subsequent infections, the CRISPR loci are transcribed and processed into guide sequences for the neutralization of the invading RNA or DNA. The CRISPR-associated protein machinery (Cas) lies at the heart of this process, yet many of the molecular details of the CRISPR/Cas system remain to be elucidated. Here, we report the first structure of Csa3, a CRISPR-associated protein from Sulfolobus solfataricus (Sso1445), which reveals a dimeric two-domain protein. The N-terminal domain is a unique variation on the dinucleotide binding domain that orchestrates dimer formation. In addition, it utilizes two conserved sequence motifs [Thr-h-Gly-Phe-(Asn/Asp)-Glu-X(4)-Arg and Leu-X(2)-Gly-h-Arg] to construct a 2-fold symmetric pocket on the dimer axis. This pocket is likely to represent a regulatory ligand-binding site. The N-terminal domain is fused to a C-terminal MarR-like winged helix-turn-helix domain that is expected to be involved in DNA recognition. Overall, the unique domain architecture of Csa3 suggests a transcriptional regulator under allosteric control of the N-terminal domain. Alternatively, Csa3 may function in a larger complex, with the conserved cleft participating in protein-protein or protein-nucleic acid interactions. A similar N-terminal domain is also identified in Csx1, a second CRISPR-associated protein family of unknown function.
Organizational Affiliation:
Thermal Biology Institute, Montana State University, Bozeman, MT 59717, USA.