Crystal Structure of the Catalytic Domain of Haspin, an Atypical Kinase Implicated in Chromatin Organization.
Villa, F., Capasso, P., Tortorici, M., Forneris, F., De Marco, A., Mattevi, A., Musacchio, A.(2009) Proc Natl Acad Sci U S A 106: 20204
- PubMed: 19918049
- DOI: https://doi.org/10.1073/pnas.0908485106
- Primary Citation of Related Structures:
2WB8 - PubMed Abstract:
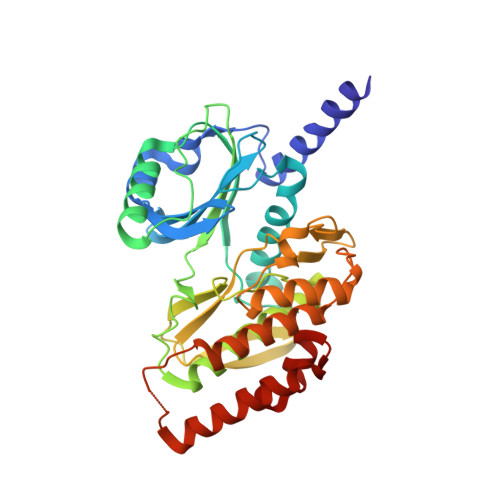
Haspin, a nuclear and chromosome-associated serine/threonine (S/T) kinase, is responsible for mitotic phosphorylation of Thr-3 of histone H3. Haspin bears recognizable similarity to the eukaryotic protein kinase (ePK) fold, but its sequence is highly divergent and there is therefore considerable interest in its structural organization. We report the 2.15-A crystal structure of the kinase domain of human Haspin. The ePK fold of Haspin contains an array of insertions and deletions. The structure illustrates how Haspin escapes the classical activation scheme of most other kinases. The alphaC helix, which bears a conserved glutamate that is essential for catalysis, adopts its final active conformation within the small lobe of the kinase. It is sandwiched between an alpha-helical insertion that precedes the kinase domain, and the activation segment, which adopts an unprecedented conformation. The activation segment, which does not contain phosphorylatable residues, packs against an unusually structured alphaEF helix. Significantly extruded from the core of the fold, it forms an extensive plateau, hosting several residues implicated in substrate binding. Overall, the structure of the Haspin kinase domain reveals an active conformation that is poised for substrate recognition and phosphorylation in the absence of external regulators.
Organizational Affiliation:
Department of Experimental Oncology, European Institute of Oncology, Via Adamello 16, 20139 Milan, Italy. fabrizio.villa@ifom-ieo-campus.it