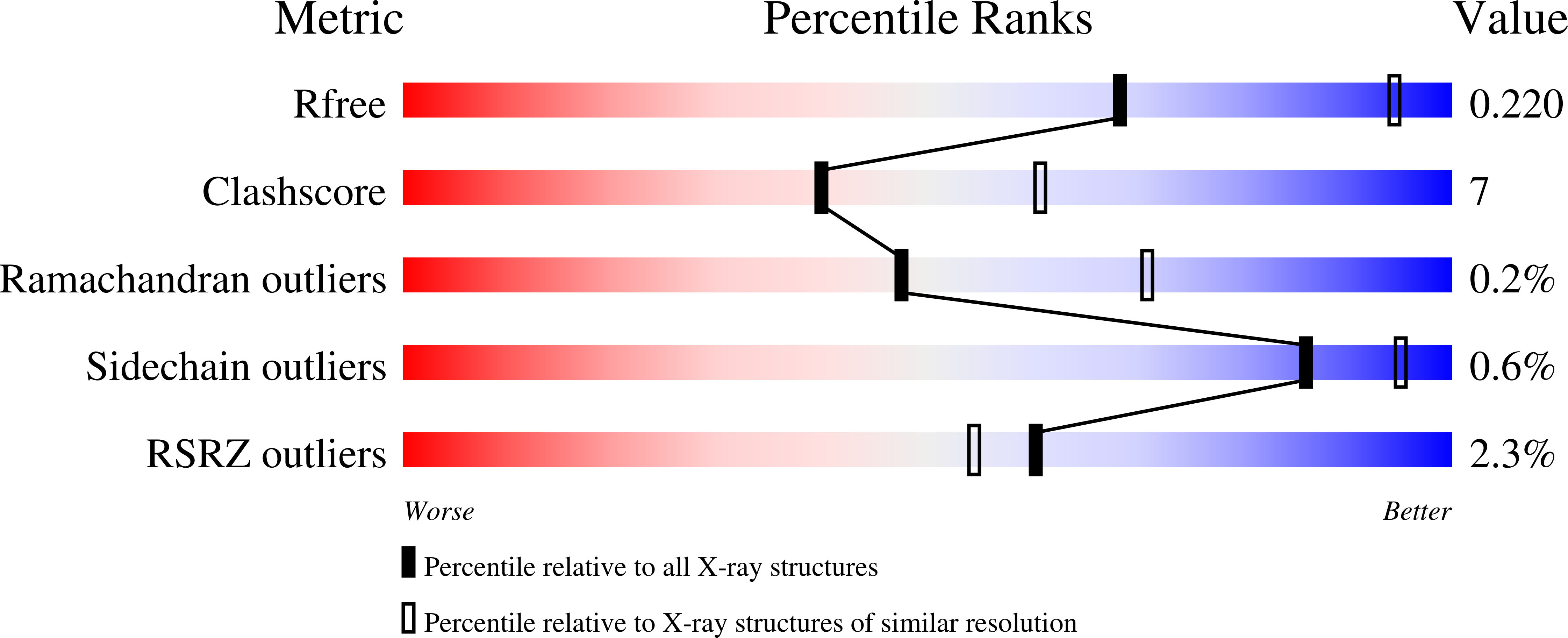
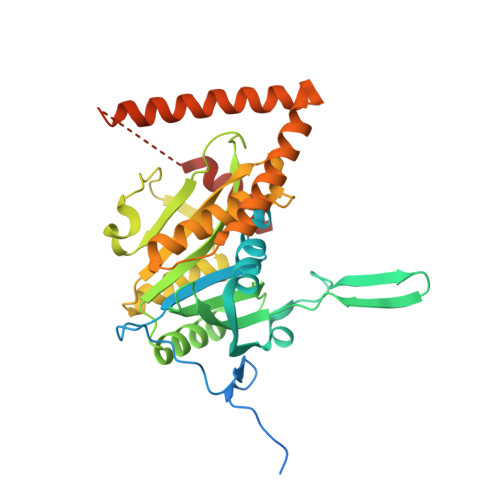
Structure of Mycobacterium Tuberculosis Rv2714, a Representative of a Duplicated Gene Family in Actinobacteria.
Grana, M., Bellinzoni, M., Miras, I., Fiex-Vandal, C., Haouz, A., Shepard, W., Buschiazzo, A., Alzari, P.M.(2009) Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 972
- PubMed: 19851001
- DOI: https://doi.org/10.1107/S1744309109035027
- Primary Citation of Related Structures:
2WAM - PubMed Abstract:
The gene Rv2714 from Mycobacterium tuberculosis, which codes for a hypothetical protein of unknown function, is a representative member of a gene family that is largely confined to the order Actinomycetales of Actinobacteria. Sequence analysis indicates the presence of two paralogous genes in most mycobacterial genomes and suggests that gene duplication was an ancient event in bacterial evolution. The crystal structure of Rv2714 has been determined at 2.6 A resolution, revealing a trimer in which the topology of the protomer core is similar to that observed in a functionally diverse set of enzymes, including purine nucleoside phosphorylases, some carboxypeptidases, bacterial peptidyl-tRNA hydrolases and even the plastidic form of an intron splicing factor. However, some structural elements, such as a beta-hairpin insertion involved in protein oligomerization and a C-terminal alpha-helical domain that serves as a lid to the putative substrate-binding (or ligand-binding) site, are only found in Rv2714 bacterial homologues and represent specific signatures of this protein family.
Organizational Affiliation:
Institut Pasteur, Unité de Biochimie Structurale, URA CNRS 2185, 25 Rue du Dr Roux, 75724 Paris, France.