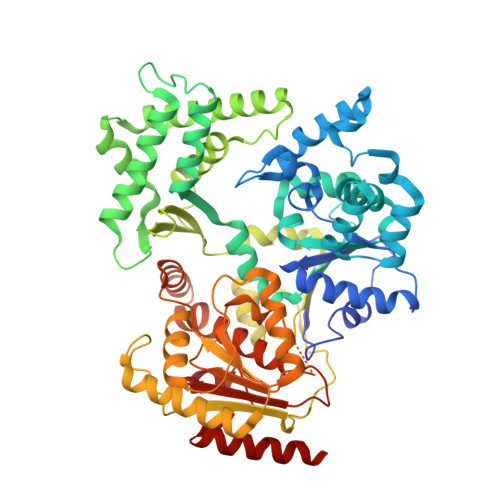
Crystal Structure of the Fes Cluster-Containing Nucleotide Excision Repair Helicase Xpd.
Wolski, S.C., Kuper, J., Haenzelmann, P., Truglio, J.J., Croteau, D.L., Van Houten, B., Kisker, C.(2008) PLoS Biol 6: E149
- PubMed: 18578568
- DOI: https://doi.org/10.1371/journal.pbio.0060149
- Primary Citation of Related Structures:
2VSF - PubMed Abstract:
DNA damage recognition by the nucleotide excision repair pathway requires an initial step identifying helical distortions in the DNA and a proofreading step verifying the presence of a lesion. This proofreading step is accomplished in eukaryotes by the TFIIH complex. The critical damage recognition component of TFIIH is the XPD protein, a DNA helicase that unwinds DNA and identifies the damage. Here, we describe the crystal structure of an archaeal XPD protein with high sequence identity to the human XPD protein that reveals how the structural helicase framework is combined with additional elements for strand separation and DNA scanning. Two RecA-like helicase domains are complemented by a 4Fe4S cluster domain, which has been implicated in damage recognition, and an alpha-helical domain. The first helicase domain together with the helical and 4Fe4S-cluster-containing domains form a central hole with a diameter sufficient in size to allow passage of a single stranded DNA. Based on our results, we suggest a model of how DNA is bound to the XPD protein, and can rationalize several of the mutations in the human XPD gene that lead to one of three severe diseases, xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy.
Organizational Affiliation:
Rudolf Virchow Center for Experimental Biomedicine, Institute for Structural Biology, University of Würzburg, Würzburg, Germany.