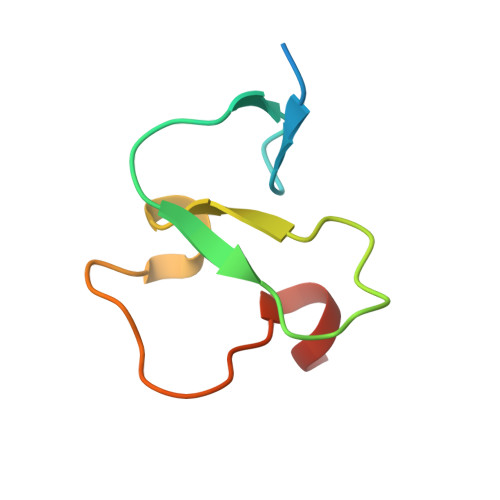
Molecular Basis of Histone H3K4Me3 Recognition by Ing4
Palacios, A., Munoz, I.G., Pantoja-Uceda, D., Marcaida, M.J., Torres, D., Martin-Garcia, J.M., Luque, I., Montoya, G., Blanco, F.J.(2008) J Biol Chem 283: 15956
- PubMed: 18381289
- DOI: https://doi.org/10.1074/jbc.M710020200
- Primary Citation of Related Structures:
2VNF - PubMed Abstract:
The inhibitors of growth (ING) family of tumor suppressors consists of five homologous proteins involved in chromatin remodeling. They form part of different acetylation and deacetylation complexes and are thought to direct them to specific regions of the chromatin, through the recognition of H3K4me3 (trimethylated K4 in the histone 3 tail) by their conserved plant homeodomain (PHD). We have determined the crystal structure of ING4-PHD bound to H3K4me3, which reveals a tight complex stabilized by numerous interactions. NMR shows that there is a reduction in the backbone mobility on the regions of the PHD that participate in the peptide binding, and binding affinities differ depending on histone tail lengths Thermodynamic analysis reveals that the discrimination in favor of methylated lysine is entropy-driven, contrary to what has been described for chromodomains. The molecular basis of H3K4me3 recognition by ING4 differs from that of ING2, which is consistent with their different affinities for methylated histone tails. These differences suggest a distinct role in transcriptional regulation for these two ING family members because of the antagonistic effect of the complexes that they recruit onto chromatin. Our results illustrate the versatility of PHD fingers as readers of the histone code.
Organizational Affiliation:
Structural Biology Unit, CIC bioGUNE, Parque Tecnológico de Bizkaia, Edificio 800, 48160 Derio, Spain.