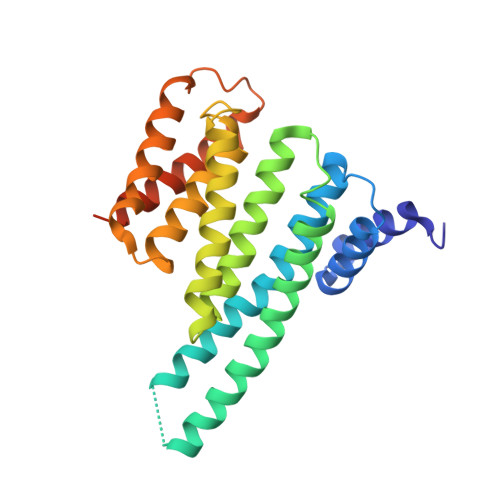
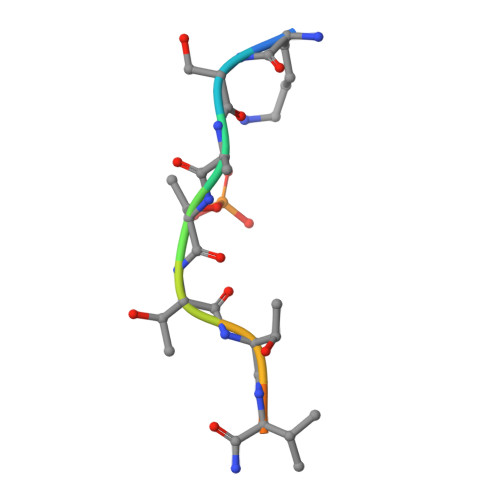
Beta2 Integrin Phosphorylation on Thr758 Acts as a Molecular Switch to Regulate 14-3-3 and Filamin Binding.
Takala, H., Nurminen, E., Nurmi, S.M., Aatonen, M., Strandin, T., Takatalo, M., Kiema, T., Gahmberg, C.G., Ylanne, J., Fagerholm, S.C.(2008) Blood 112: 1853
- PubMed: 18550856
- DOI: https://doi.org/10.1182/blood-2007-12-127795
- Primary Citation of Related Structures:
2JF1, 2V7D - PubMed Abstract:
Leukocyte integrins of the beta2 family are essential for immune cell-cell adhesion. In activated cells, beta2 integrins are phosphorylated on the cytoplasmic Thr758, leading to 14-3-3 protein recruitment to the beta2 integrin. The mutation of this phosphorylation site impairs cell adhesion, actin reorganization, and cell spreading. Thr758 is contained in a Thr triplet of beta2 that also mediates binding to filamin. Here, we investigated the binding of filamin, talin, and 14-3-3 proteins to phosphorylated and unphosphorylated beta2 integrins by biochemical methods and x-ray crystallography. 14-3-3 proteins bound only to the phosphorylated integrin cytoplasmic peptide, with a high affinity (K(d), 261 nM), whereas filamin bound only the unphosphorylated integrin cytoplasmic peptide (K(d), 0.5 mM). Phosphorylation did not regulate talin binding to beta2 directly, but 14-3-3 was able to outcompete talin for the binding to phosphorylated beta2 integrin. X-ray crystallographic data clearly explained how phosphorylation eliminated filamin binding and induced 14-3-3 protein binding. Filamin knockdown in T cells led to an increase in stimulated cell adhesion to ICAM-1-coated surfaces. Our results suggest that the phosphorylation of beta2 integrins on Thr758 acts as a molecular switch to inhibit filamin binding and allow 14-3-3 protein binding to the integrin cytoplasmic domain, thereby modulating T-cell adhesion.
Organizational Affiliation:
Department of Biological and Environmental Science and Nanoscience Center, University of Jyväskylä, Jyväskylä, Finland.