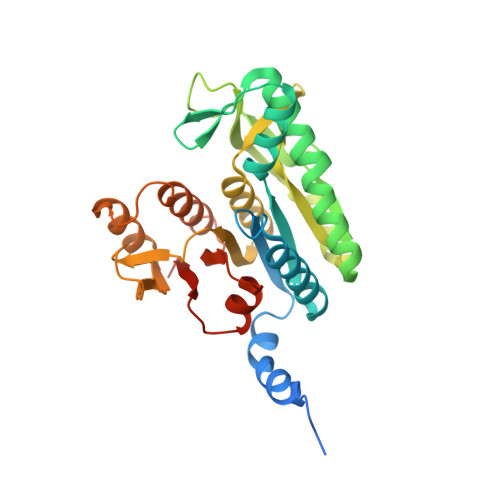
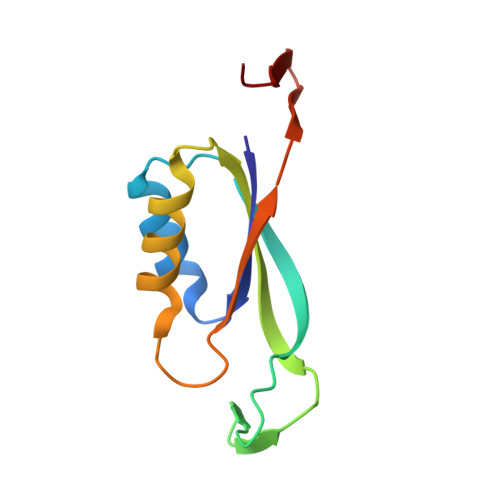
The Crystal Structure of the Complex of Pii and Acetylglutamate Kinase Reveals How Pii Controls the Storage of Nitrogen as Arginine.
Llacer, J.L., Contreras, A., Forchhammer, K., Marco-Marin, C., Gil-Ortiz, F., Maldonado, R., Fita, I., Rubio, V.(2007) Proc Natl Acad Sci U S A 104: 17644
- PubMed: 17959776
- DOI: https://doi.org/10.1073/pnas.0705987104
- Primary Citation of Related Structures:
2JJ4, 2V5H - PubMed Abstract:
Photosynthetic organisms can store nitrogen by synthesizing arginine, and, therefore, feedback inhibition of arginine synthesis must be relieved in these organisms when nitrogen is abundant. This relief is accomplished by the binding of the PII signal transduction protein to acetylglutamate kinase (NAGK), the controlling enzyme of arginine synthesis. Here, we describe the crystal structure of the complex between NAGK and PII of Synechococcus elongatus, at 2.75-A resolution. We prove the physiological relevance of the observed interactions by site-directed mutagenesis and functional studies. The complex consists of two polar PII trimers sandwiching one ring-like hexameric NAGK (a trimer of dimers) with the threefold axes of these molecules aligned. The binding of PII favors a narrow ring conformation of the NAGK hexamer that is associated with arginine sites having low affinity for this inhibitor. Each PII subunit contacts one NAGK subunit only. The contacts map in the inner circumference of the NAGK ring and involve two surfaces of the PII subunit. One surface is on the PII body and interacts with the C-domain of the NAGK subunit, helping widen the arginine site found on the other side of this domain. The other surface is at the distal region of a protruding large loop (T-loop) that presents a novel compact shape. This loop is inserted in the interdomain crevice of the NAGK subunit, contacting mainly the N-domain, and playing key roles in anchoring PII on NAGK, in activating NAGK, and in complex formation regulation by MgATP, ADP, 2-oxoglutarate, and by phosphorylation of serine-49.
Organizational Affiliation:
Instituto de Biomedicina de Valencia-Consejo Superior de Investigaciones Cientificas and Centro de Investigación Biomédica en Red de Enfermedades Raras, Jaime Roig 11, 46010 Valencia, Spain.