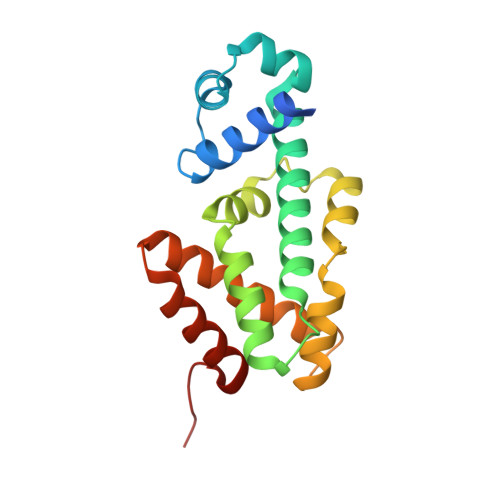
Structural Plasticity and Distinct Drug-Binding Modes of Lfrr, a Mycobacterial Efflux Pump Regulator.
Bellinzoni, M., Buroni, S., Schaeffer, F., Riccardi, G., De Rossi, E., Alzari, P.M.(2009) J Bacteriol 191: 7531
- PubMed: 19820093
- DOI: https://doi.org/10.1128/JB.00631-09
- Primary Citation of Related Structures:
2V57, 2WGB - PubMed Abstract:
The TetR-like transcriptional repressor LfrR controls the expression of the gene encoding the Mycobacterium smegmatis efflux pump LfrA, which actively extrudes fluoroquinolones, cationic dyes, and anthracyclines from the cell and promotes intrinsic antibiotic resistance. The crystal structure of the apoprotein form of the repressor reveals a structurally asymmetric homodimer exhibiting local unfolding and a blocked drug-binding site, emphasizing the significant conformational plasticity of the protein necessary for DNA and multidrug recognition. Crystallographic and calorimetric studies of LfrR-drug complexes further confirm the intrinsic flexibility of the homodimer, which provides a dynamic mechanism to broaden multidrug binding specificity and may be a general property of transcriptional repressors regulating microbial efflux pump expression.
Organizational Affiliation:
Institut Pasteur, Unité de Biochimie Structurale and CNRS URA 2185, Institut Pasteur, 75724 Paris Cedex 15, France.