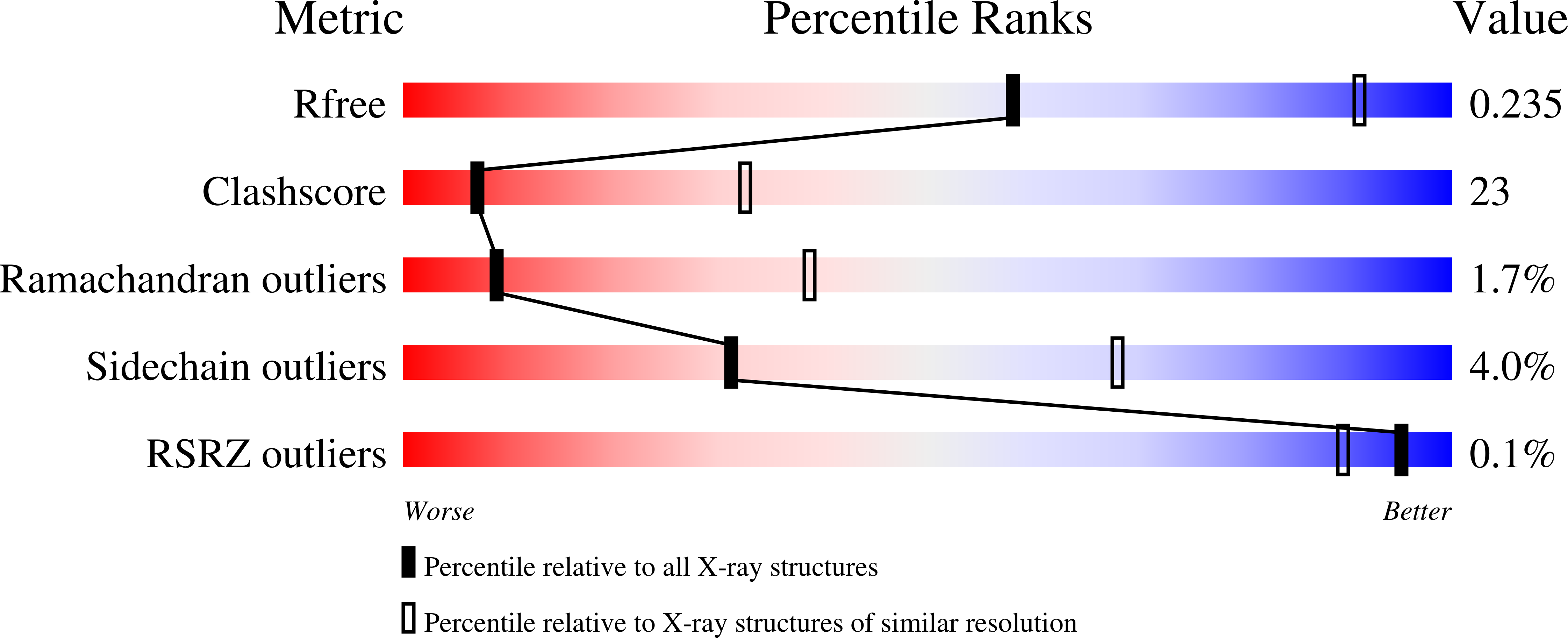
Mycobacterium Tuberculosis Cyp130: Crystal Structure, Biophysical Characterization, and Interactions with Antifungal Azole Drugs
Ouellet, H., Podust, L.M., Ortiz de Montellano, P.R.(2008) J Biol Chem 283: 5069
- PubMed: 18089574
- DOI: https://doi.org/10.1074/jbc.M708734200
- Primary Citation of Related Structures:
2UUQ, 2UVN - PubMed Abstract:
CYP130 is one of the 20 Mycobacterium tuberculosis cytochrome P450 enzymes, only two of which, CYP51 and CYP121, have so far been studied as individually expressed proteins. Here we characterize a third heterologously expressed M. tuberculosis cytochrome P450, CYP130, by UV-visible spectroscopy, isothermal titration calorimetry, and x-ray crystallography, including determination of the crystal structures of ligand-free and econazole-bound CYP130 at a resolution of 1.46 and 3.0A(,) respectively. Ligand-free CYP130 crystallizes in an "open" conformation as a monomer, whereas the econazole-bound form crystallizes in a "closed" conformation as a dimer. Conformational changes enabling the "open-closed" transition involve repositioning of the BC-loop and the F and G helices that envelop the inhibitor in the binding site and reshape the protein surface. Crystal structure analysis shows that the portion of the BC-loop relocates as much as 18A between the open and closed conformations. Binding of econazole to CYP130 involves a conformational change and is mediated by both a set of hydrophobic interactions with amino acid residues in the active site and coordination of the heme iron. CYP130 also binds miconazole with virtually the same binding affinity as econazole and clotrimazole and ketoconazole with somewhat lower affinities, which makes it a plausible target for this class of therapeutic drugs. Overall, binding of the azole inhibitors is a sequential two-step, entropy-driven endothermic process. Binding of econazole and clotrimazole exhibits positive cooperativity that may reflect a propensity of CYP130 to associate into a dimeric structure.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco, California 94158, USA.