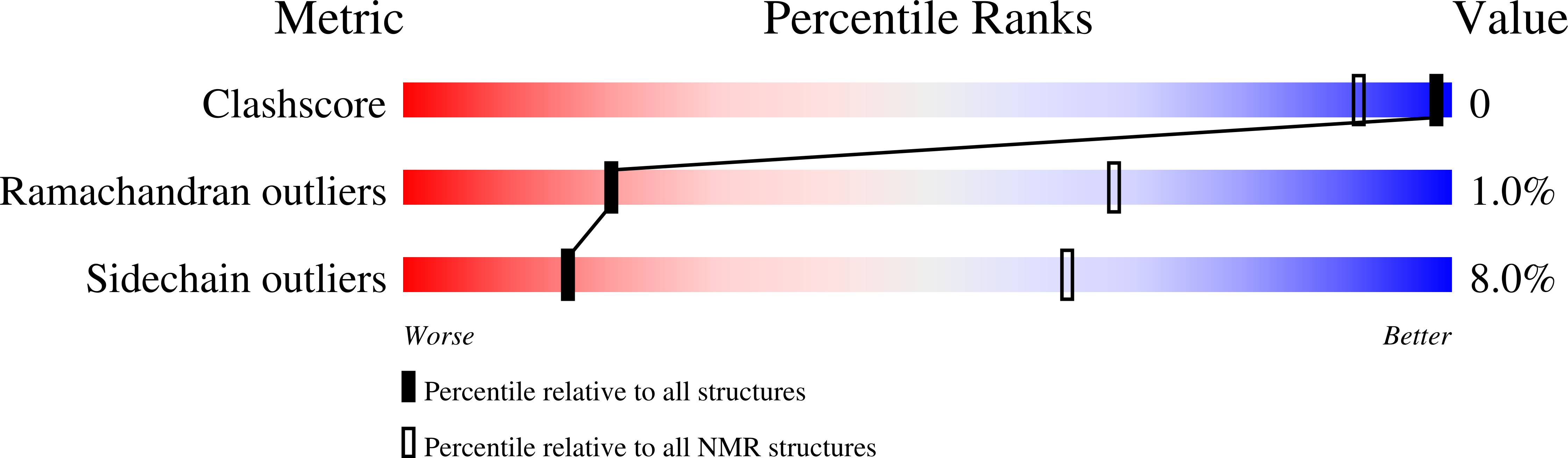Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies
Furuita, K., Jee, J., Fukada, H., Mishima, M., Kojima, C.(2010) J Biol Chem 285: 12961-12970
- PubMed: 20178991
- DOI: https://doi.org/10.1074/jbc.M109.082602
- Primary Citation of Related Structures:
2RR3 - PubMed Abstract:
Oxysterol-binding protein (OSBP), a cytosolic receptor of cholesterol and oxysterols, is recruited to the endoplasmic reticulum by binding to the cytoplasmic major sperm protein (MSP) domain of integral endoplasmic reticulum protein VAMP-associated protein-A (VAP-A), a process essential for the stimulation of sphingomyelin synthesis by 25-hydroxycholesterol. To delineate the interaction mechanism between VAP-A and OSBP, we determined the complex structure between the VAP-A MSP domain (VAP-A(MSP)) and the OSBP fragment containing a VAP-A binding motif FFAT (OSBP(F)) by NMR. This solution structure explained that five of six conserved residues in the FFAT motif are required for the stable complex formation, and three of five, including three critical intermolecular electrostatic interactions, were not explained before. By combining NMR relaxation and titration, isothermal titration calorimetry, and mutagenesis experiments with structural information, we further elucidated the detailed roles of the FFAT motif and underlying motions of VAP-A(MSP), OSBP(F), and the complex. Our results show that OSBP(F) is disordered in the free state, and VAP-A(MSP) and OSBP(F) form a final complex by means of intermediates, where electrostatic interactions through acidic residues, including an acid patch preceding the FFAT motif, probably play a collective role. Additionally, we report that the mutation that causes the familial motor neuron disease decreases the stability of the MSP domain.
Organizational Affiliation:
Graduate School of Biological Sciences, Nara Institute of Science and Technology, Nara 630-0192, Japan.