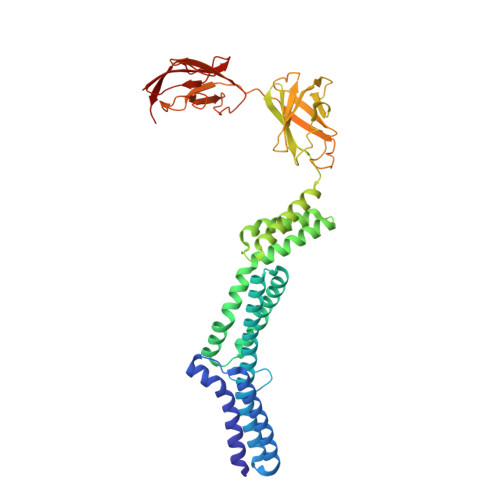
The structure and binding behavior of the bacterial cell surface layer protein SbsC.
Pavkov, T., Egelseer, E.M., Tesarz, M., Svergun, D.I., Sleytr, U.B., Keller, W.(2008) Structure 16: 1226-1237
- PubMed: 18682224
- DOI: https://doi.org/10.1016/j.str.2008.05.012
- Primary Citation of Related Structures:
2RA1 - PubMed Abstract:
Surface layers (S-layers) comprise the outermost cell envelope component of most archaea and many bacteria. Here we present the structure of the bacterial S-layer protein SbsC from Geobacillus stearothermophilus, showing a very elongated and flexible molecule, with strong and specific binding to the secondary cell wall polymer (SCWP). The crystal structure of rSbsC((31-844)) revealed a novel fold, consisting of six separate domains, which are connected by short flexible linkers. The N-terminal domain exhibits positively charged residues regularly spaced along the putative ligand binding site matching the distance of the negative charges on the extended SCWP. Upon SCWP binding, a considerable stabilization of the N-terminal domain occurs. These findings provide insight into the processes of S-layer attachment to the underlying cell wall and self-assembly, and also accommodate the observed mechanical strength, the polarity of the S-layer, and the pronounced requirement for surface flexibility inherent to cell growth and division.
Organizational Affiliation:
Institute of Molecular Biosciences, Structural Biology, University of Graz, Humboldtsrasse 50/3, 8010 Graz, Austria.