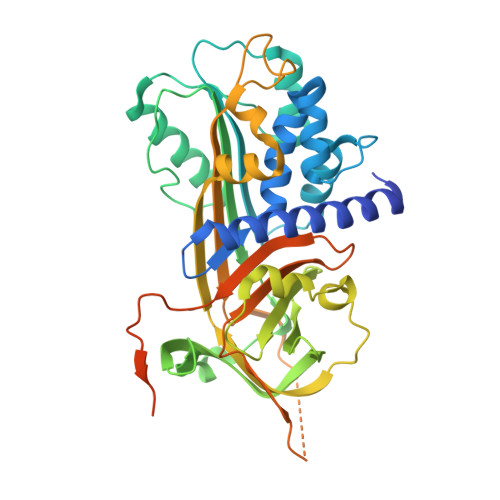
X-ray crystal structure of the fibrinolysis inhibitor {alpha}2-antiplasmin
Law, R.H.P., Sofian, T., Kan, W.T., Horvath, A.J., Hitchen, C.R., Langendorf, C.G., Buckle, A.M., Whisstock, J.C., Coughlin, P.B.(2008) Blood 111: 2049-2052
- PubMed: 18063751
- DOI: https://doi.org/10.1182/blood-2007-09-114215
- Primary Citation of Related Structures:
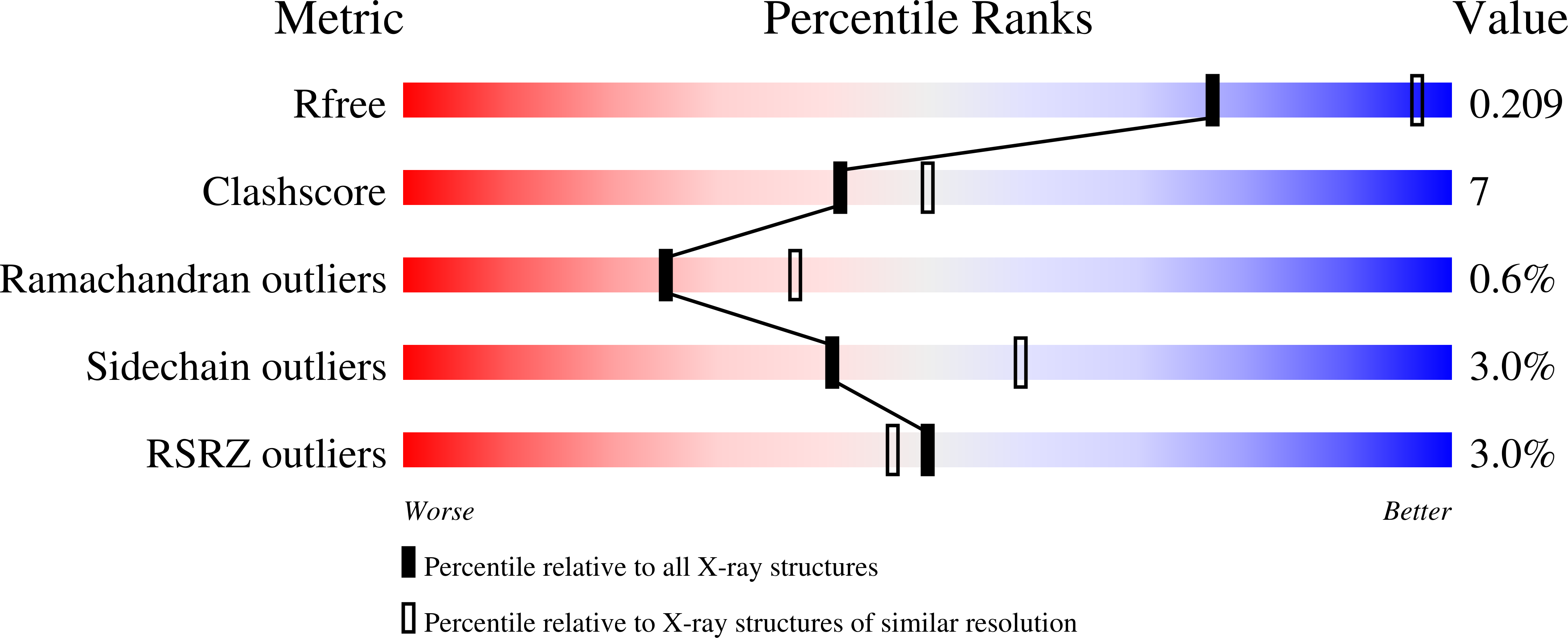
2R9Y - PubMed Abstract:
The serpin alpha(2)-antiplasmin (SERPINF2) is the principal inhibitor of plasmin and inhibits fibrinolysis. Accordingly, alpha(2)-antiplasmin deficiency in humans results in uncontrolled fibrinolysis and a bleeding disorder. alpha(2)-antiplasmin is an unusual serpin, in that it contains extensive N- and C-terminal sequences flanking the serpin domain. The N-terminal sequence is crosslinked to fibrin by factor XIIIa, whereas the C-terminal region mediates the initial interaction with plasmin. To understand how this may happen, we have determined the 2.65A X-ray crystal structure of an N-terminal truncated murine alpha(2)-antiplasmin. The structure reveals that part of the C-terminal sequence is tightly associated with the body of the serpin. This would be anticipated to position the flexible plasmin-binding portion of the C-terminus in close proximity to the serpin Reactive Center Loop where it may act as a template to accelerate serpin/protease interactions.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia