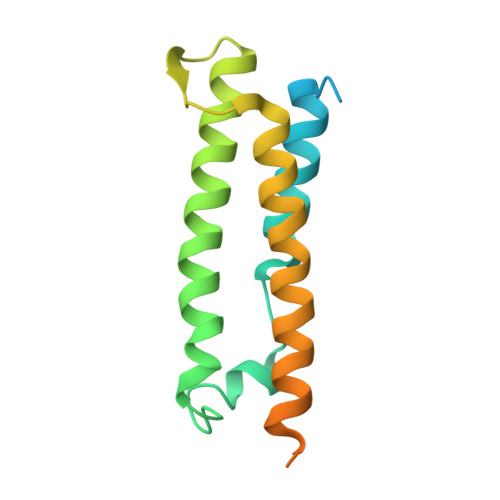
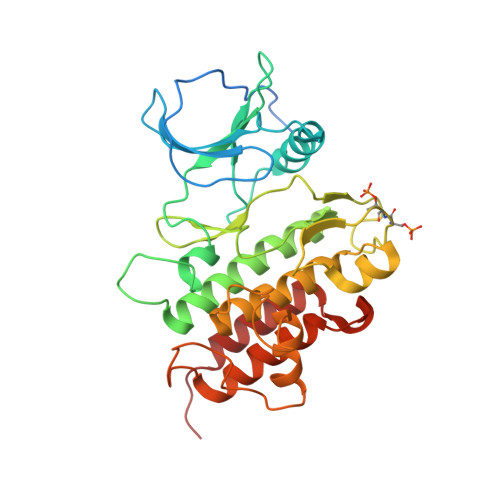
The structural basis for activation of plant immunity by bacterial effector protein AvrPto
Xing, W., Zou, Y., Liu, Q., Liu, J., Luo, X., Huang, Q., Chen, S., Zhu, L., Bi, R., Hao, Q., Wu, J.W., Zhou, J.M., Chai, J.(2007) Nature 449: 243-247
- PubMed: 17694048
- DOI: https://doi.org/10.1038/nature06109
- Primary Citation of Related Structures:
2QKW - PubMed Abstract:
Pathogenic microbes use effectors to enhance susceptibility in host plants. However, plants have evolved a sophisticated immune system to detect these effectors using cognate disease resistance proteins, a recognition that is highly specific, often elicits rapid and localized cell death, known as a hypersensitive response, and thus potentially limits pathogen growth. Despite numerous genetic and biochemical studies on the interactions between pathogen effector proteins and plant resistance proteins, the structural bases for such interactions remain elusive. The direct interaction between the tomato protein kinase Pto and the Pseudomonas syringae effector protein AvrPto is known to trigger disease resistance and programmed cell death through the nucleotide-binding site/leucine-rich repeat (NBS-LRR) class of disease resistance protein Prf. Here we present the crystal structure of an AvrPto-Pto complex. Contrary to the widely held hypothesis that AvrPto activates Pto kinase activity, our structural and biochemical analyses demonstrated that AvrPto is an inhibitor of Pto kinase in vitro. The AvrPto-Pto interaction is mediated by the phosphorylation-stabilized P+1 loop and a second loop in Pto, both of which negatively regulate the Prf-mediated defences in the absence of AvrPto in tomato plants. Together, our results show that AvrPto derepresses host defences by interacting with the two defence-inhibition loops of Pto.
Organizational Affiliation:
National Institute of Biological Sciences, No. 7 Science Park Road, Beijing 102206, China.