The Structural Basis for Unidirectional Rotation of Thermoalkaliphilic F(1)-ATPase.
Stocker, A., Keis, S., Vonck, J., Cook, G.M., Dimroth, P.(2007) Structure 15: 904-914
- PubMed: 17697996
- DOI: https://doi.org/10.1016/j.str.2007.06.009
- Primary Citation of Related Structures:
2QE7 - PubMed Abstract:
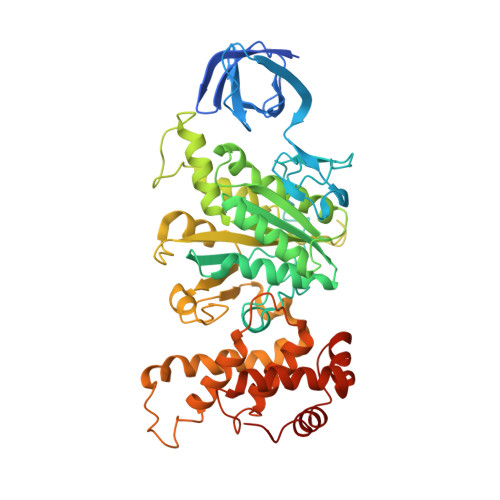
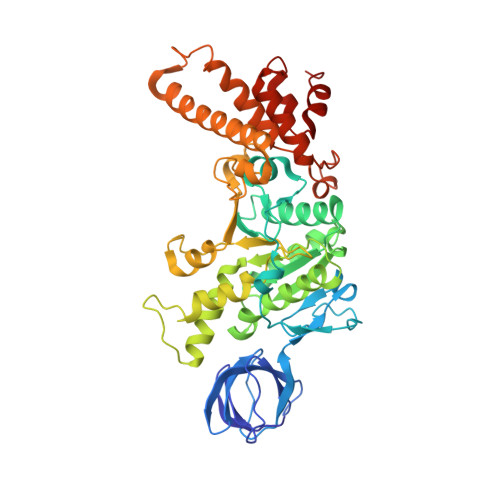
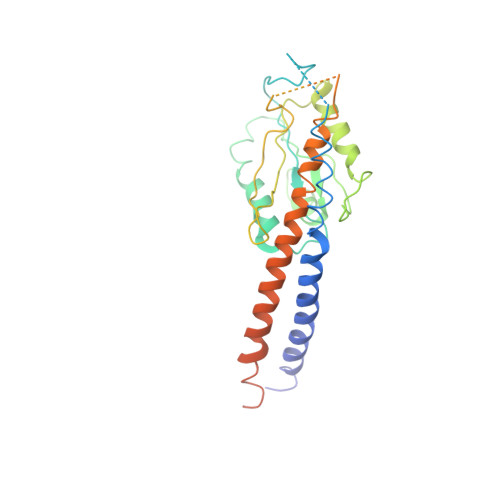
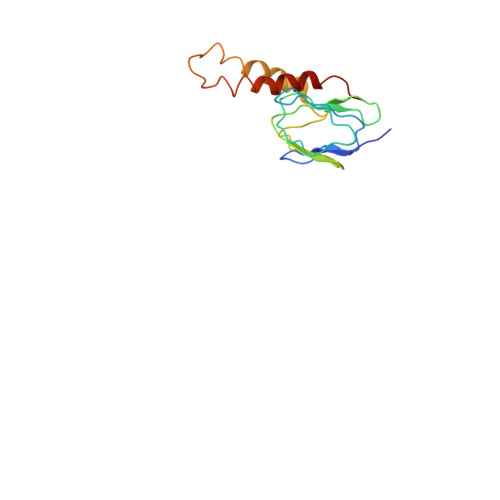
The ATP synthase of the thermoalkaliphilic Bacillus sp. TA2.A1 operates exclusively in ATP synthesis direction. In the crystal structure of the nucleotide-free alpha(3)beta(3)gamma epsilon subcomplex (TA2F(1)) at 3.1 A resolution, all three beta subunits adopt the open beta(E) conformation. The structure shows salt bridges between the helix-turn-helix motif of the C-terminal domain of the beta(E) subunit (residues Asp372 and Asp375) and the N-terminal helix of the gamma subunit (residues Arg9 and Arg10). These electrostatic forces pull the gamma shaft out of the rotational center and impede rotation through steric interference with the beta(E) subunit. Replacement of Arg9 and Arg10 with glutamines eliminates the salt bridges and results in an activation of ATP hydrolysis activity, suggesting that these salt bridges prevent the native enzyme from rotating in ATP hydrolysis direction. A similar bending of the gamma shaft as in the TA2F(1) structure was observed by single-particle analysis of the TA2F(1)F(o) holoenzyme.
Organizational Affiliation:
Institute of Microbiology ETH Zürich, ETH Hönggerberg, Wolfgang-Pauli-Strasse 10, CH-8093 Zürich, Switzerland.