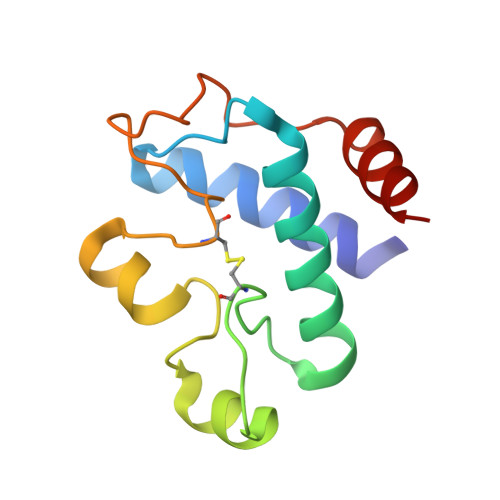
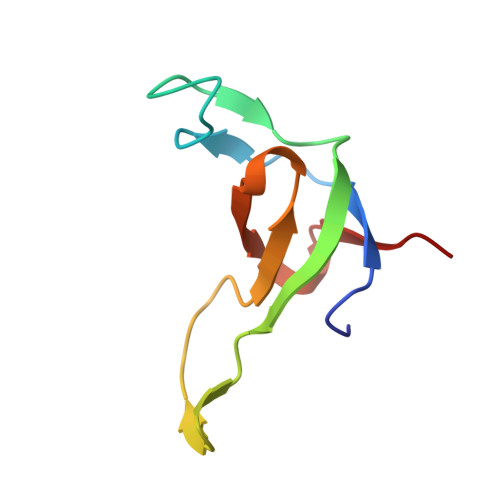
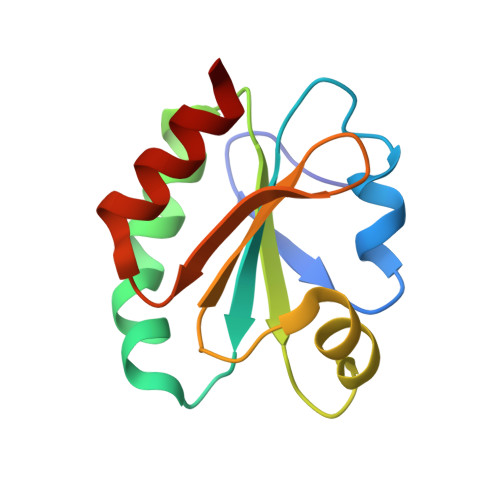
Structural snapshots along the reaction pathway of ferredoxin-thioredoxin reductase.
Dai, S., Friemann, R., Glauser, D.A., Bourquin, F., Manieri, W., Schurmann, P., Eklund, H.(2007) Nature 448: 92-96
- PubMed: 17611542
- DOI: https://doi.org/10.1038/nature05937
- Primary Citation of Related Structures:
2PU9, 2PUK, 2PUO, 2PVD, 2PVG, 2PVO - PubMed Abstract:
Oxygen-evolving photosynthetic organisms regulate carbon metabolism through a light-dependent redox signalling pathway. Electrons are shuttled from photosystem I by means of ferredoxin (Fdx) to ferredoxin-thioredoxin reductase (FTR), which catalyses the two-electron-reduction of chloroplast thioredoxins (Trxs). These modify target enzyme activities by reduction, regulating carbon flow. FTR is unique in its use of a [4Fe-4S] cluster and a proximal disulphide bridge in the conversion of a light signal into a thiol signal. We determined the structures of FTR in both its one- and its two-electron-reduced intermediate states and of four complexes in the pathway, including the ternary Fdx-FTR-Trx complex. Here we show that, in the first complex (Fdx-FTR) of the pathway, the Fdx [2Fe-2S] cluster is positioned suitably for electron transfer to the FTR [4Fe-4S] centre. After the transfer of one electron, an intermediate is formed in which one sulphur atom of the FTR active site is free to attack a disulphide bridge in Trx and the other sulphur atom forms a fifth ligand for an iron atom in the FTR [4Fe-4S] centre--a unique structure in biology. Fdx then delivers a second electron that cleaves the FTR-Trx heterodisulphide bond, which occurs in the Fdx-FTR-Trx complex. In this structure, the redox centres of the three proteins are aligned to maximize the efficiency of electron transfer from the Fdx [2Fe-2S] cluster to the active-site disulphide of Trxs. These results provide a structural framework for understanding the mechanism of disulphide reduction by an iron-sulphur enzyme and describe previously unknown interaction networks for both Fdx and Trx (refs 4-6).
Organizational Affiliation:
Howard Hughes Medical Institute, Integrated Department of Immunology, National Jewish Medical and Research Center & University of Colorado Health Sciences Center, 1400 Jackson Street, Denver, Colorado 80206, USA. dais@njc.org