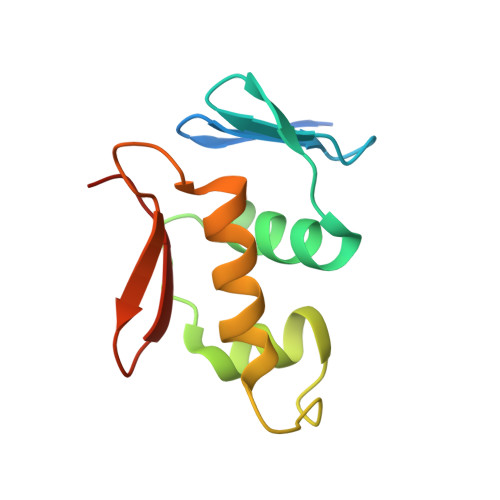
Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis
Wang, S., Engohang-Ndong, J., Smith, I.(2007) Biochemistry 46: 14751-14761
- PubMed: 18052041
- DOI: https://doi.org/10.1021/bi700970a
- Primary Citation of Related Structures:
2PMU - PubMed Abstract:
The PhoP-PhoR two-component signaling system from Mycobacterium tuberculosis is essential for the virulence of the tubercle bacillus. The response regulator, PhoP, regulates expression of over 110 genes. In order to elucidate the regulatory mechanism of PhoP, we determined the crystal structure of its DNA-binding domain (PhoPC). PhoPC exhibits a typical fold of the winged helix-turn-helix subfamily of response regulators. The structure starts with a four-stranded antiparallel beta-sheet, followed by a three-helical bundle of alpha-helices, and then a C-terminal beta-hairpin, which together with a short beta-strand between the first and second helices forms a three-stranded antiparallel beta-sheet. Structural elements are packed through a hydrophobic core, with the first helix providing a scaffold for the rest of the domain to pack. The second and third helices and the long, flexible loop between them form the helix-turn-helix motif, with the third helix being the recognition helix. The C-terminal beta-hairpin turn forms the wing motif. The molecular surfaces around the recognition helix and the wing residues show strong positive electrostatic potential, consistent with their roles in DNA binding and nucleotide sequence recognition. The crystal packing of PhoPC gives a hexamer ring, with neighboring molecules interacting in a head-to-tail fashion. This packing interface suggests that PhoPC could bind DNA in a tandem association. However, this mode of DNA binding is likely to be nonspecific because the recognition helix is partially blocked and would be prevented from inserting into the major groove of DNA. Detailed structural analysis and implications with respect to DNA binding are discussed.
Organizational Affiliation:
Public Health Research Institute, New Jersey Medical School, University of Medicine and Dentistry of New Jersey, Newark, NJ 07103, USA. wang16@umdnj.edu