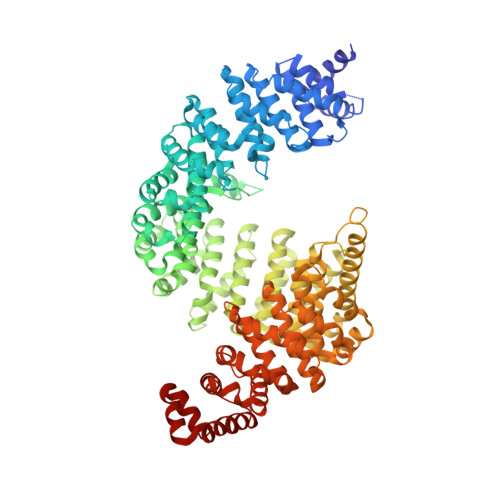
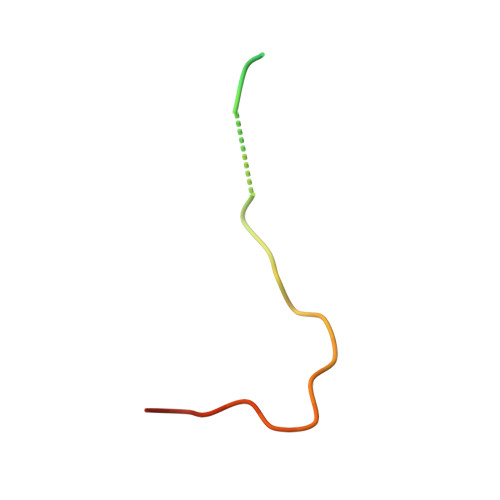
Structure-based design of a pathway-specific nuclear import inhibitor.
Cansizoglu, A.E., Lee, B.J., Zhang, Z.C., Fontoura, B.M., Chook, Y.M.(2007) Nat Struct Mol Biol 14: 452-454
- PubMed: 17435768
- DOI: https://doi.org/10.1038/nsmb1229
- Primary Citation of Related Structures:
2OT8 - PubMed Abstract:
Kapbeta2 (also called transportin) recognizes PY nuclear localization signal (NLS), a new class of NLS with a R/H/Kx((2-5))PY motif. Here we show that Kapbeta2 complexes containing hydrophobic and basic PY-NLSs, as classified by the composition of an additional N-terminal motif, converge in structure only at consensus motifs, which explains ligand diversity. On the basis of these data and complementary biochemical analyses, we designed a Kapbeta2-specific nuclear import inhibitor, M9M.
Organizational Affiliation:
Department of Pharmacology, University of Texas Southwestern Medical Center at Dallas, 6001 Forest Park, Dallas, Texas 75390-9041, USA.