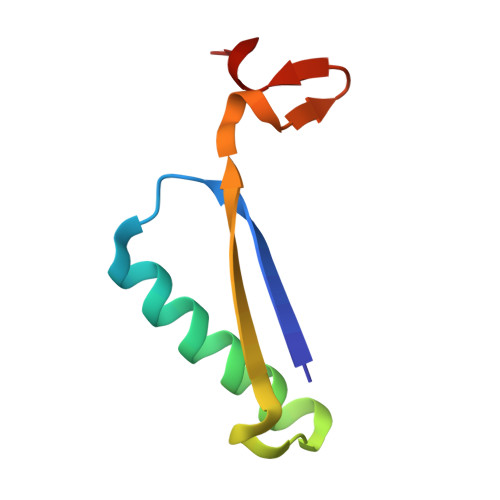
The Crystal Structure of YwhB, a 4-Oxalocrotonate Tautomerase Homologue from Bacillus subtilis: The Structural Basis for Catalysis, Inhibition, and Reaction Stereoselectivity
Hackert, M.L., Whitman, C.P., Almrud, J.J., Dasgupta, R., Wang, S.C., Johnson, W.H.To be published.