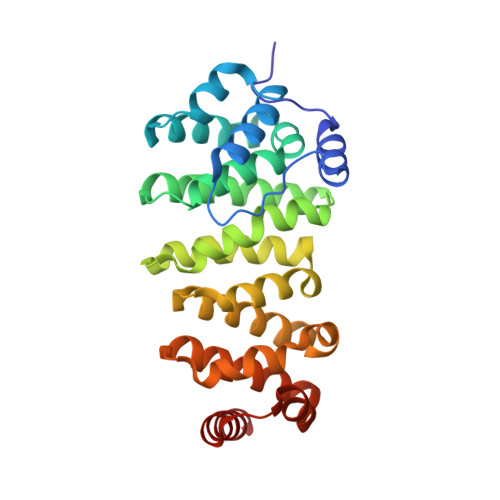
Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding.
Al-Bassam, J., Larsen, N.A., Hyman, A.A., Harrison, S.C.(2007) Structure 15: 355-362
- PubMed: 17355870
- DOI: https://doi.org/10.1016/j.str.2007.01.012
- Primary Citation of Related Structures:
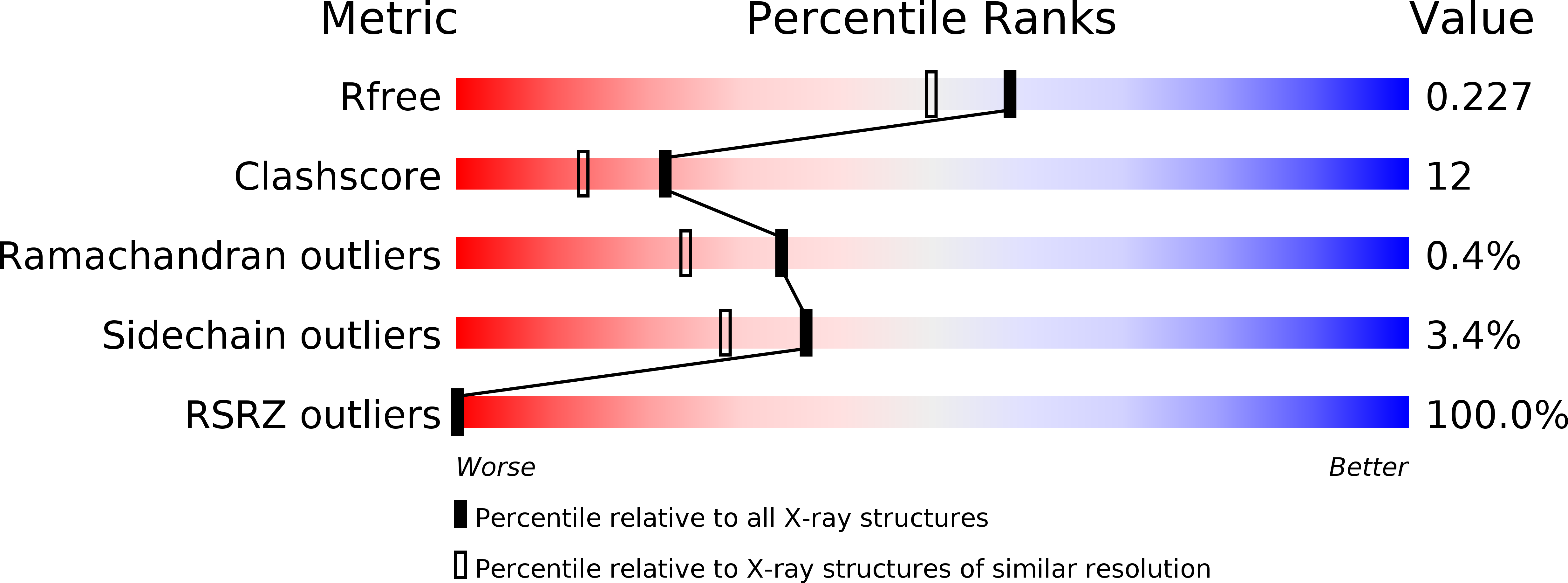
2OF3 - PubMed Abstract:
Members of the XMAP215/Dis1 family of microtubule-associated proteins (MAPs) are essential for microtubule growth. MAPs in this family contain several 250 residue repeats, called TOG domains, which are thought to bind tubulin dimers and promote microtubule polymerization. We have determined the crystal structure of a single TOG domain from the Caenorhabditis elegans homolog, Zyg9, to 1.9 A resolution, and from it we describe a structural blueprint for TOG domains. These domains are flat, paddle-like structures, composed of six HEAT-repeat elements stacked side by side. The two wide faces of the paddle contain the HEAT-repeat helices, and the two narrow faces, the intra- and inter-HEAT repeat turns. Solvent-exposed residues in the intrarepeat turns are conserved, both within a particular protein and across the XMAP215/Dis1 family. Mutation of some of these residues in the TOG1 domain from the budding yeast homolog, Stu2p, shows that this face indeed participates in the tubulin contact.
Organizational Affiliation:
Jack and Eileen Connors Laboratory of Structural Biology , Harvard Medical School, Boston, MA 02115, USA.