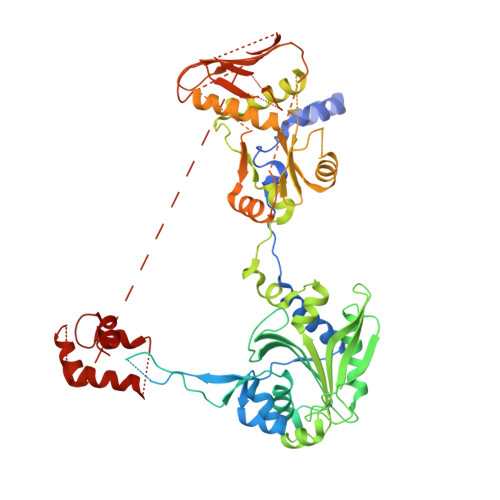
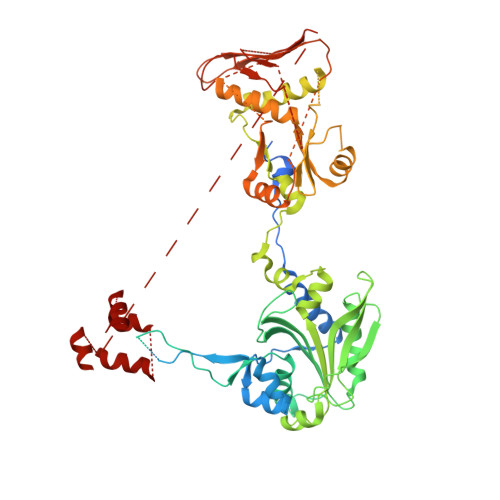
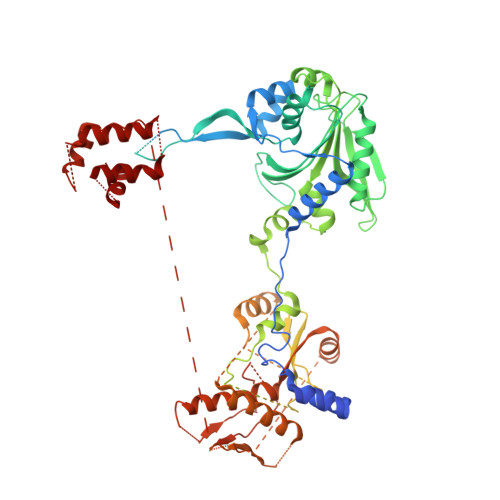
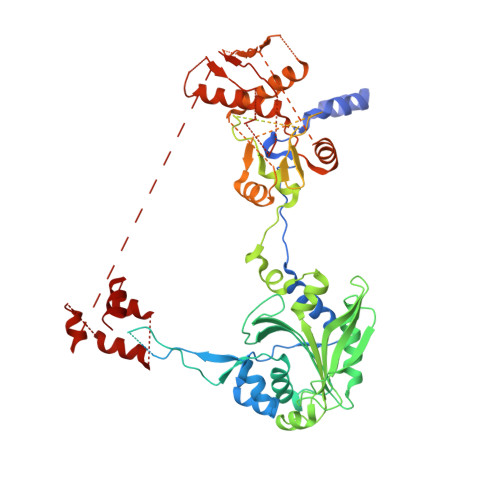
Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Kamtekar, S., Hohn, M.J., Park, H.S., Schnitzbauer, M., Sauerwald, A., Soll, D., Steitz, T.A.(2007) Proc Natl Acad Sci U S A 104: 2620-2625
- PubMed: 17301225
- DOI: https://doi.org/10.1073/pnas.0611504104
- Primary Citation of Related Structures:
2ODR - PubMed Abstract:
A number of archaeal organisms generate Cys-tRNA(Cys) in a two-step pathway, first charging phosphoserine (Sep) onto tRNA(Cys) and subsequently converting it to Cys-tRNA(Cys). We have determined, at 3.2-A resolution, the structure of the Methanococcus maripaludis phosphoseryl-tRNA synthetase (SepRS), which catalyzes the first step of this pathway. The structure shows that SepRS is a class II, alpha(4) synthetase whose quaternary structure arrangement of subunits closely resembles that of the heterotetrameric (alphabeta)(2) phenylalanyl-tRNA synthetase (PheRS). Homology modeling of a tRNA complex indicates that, in contrast to PheRS, a single monomer in the SepRS tetramer may recognize both the acceptor terminus and anticodon of a tRNA substrate. Using a complex with tungstate as a marker for the position of the phosphate moiety of Sep, we suggest that SepRS and PheRS bind their respective amino acid substrates in dissimilar orientations by using different residues.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry and Chemistry and Howard Hughes Medical Institute, Yale University, New Haven, CT 06520-8114, USA.