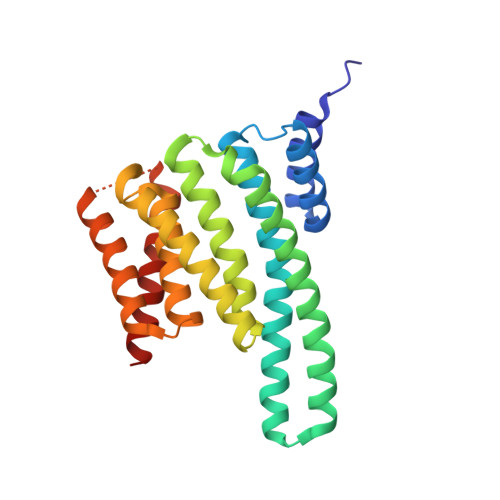
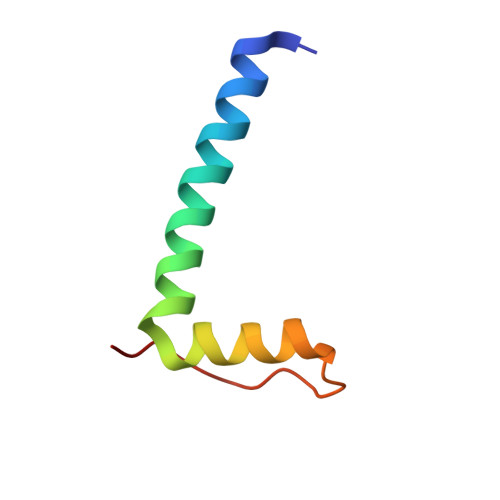
Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy
Ottmann, C., Marco, S., Jaspert, N., Marcon, C., Schauer, N., Weyand, M., Vandermeeren, C., Duby, G., Boutry, M., Wittinghofer, A., Rigaud, J.L., Oecking, C.(2007) Mol Cell 25: 427-440
- PubMed: 17289589
- DOI: https://doi.org/10.1016/j.molcel.2006.12.017
- Primary Citation of Related Structures:
2O98 - PubMed Abstract:
Regulatory 14-3-3 proteins activate the plant plasma membrane H(+)-ATPase by binding to its C-terminal autoinhibitory domain. This interaction requires phosphorylation of a C-terminal, mode III, recognition motif as well as an adjacent span of approximately 50 amino acids. Here we report the X-ray crystal structure of 14-3-3 in complex with the entire binding motif, revealing a previously unidentified mode of interaction. A 14-3-3 dimer simultaneously binds two H(+)-ATPase peptides, each of which forms a loop within the typical 14-3-3 binding groove and therefore exits from the center of the dimer. Several H(+)-ATPase mutants support this structure determination. Accordingly, 14-3-3 binding could result in H(+)-ATPase oligomerization. Indeed, by using single-particle electron cryomicroscopy, the 3D reconstruction of the purified H(+)-ATPase/14-3-3 complex demonstrates a hexameric arrangement. Fitting of 14-3-3 and H(+)-ATPase atomic structures into the 3D reconstruction map suggests the spatial arrangement of the holocomplex.
Organizational Affiliation:
Zentrum für Molekularbiologie der Pflanzen, Pflanzenphysiologie, Universität Tübingen, Auf der Morgenstelle 5, 72076 Tübingen, Germany.