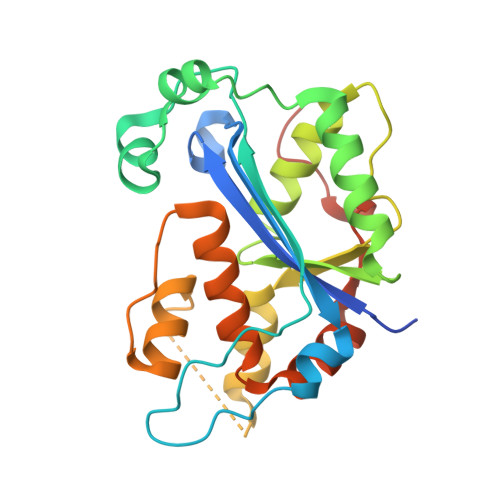
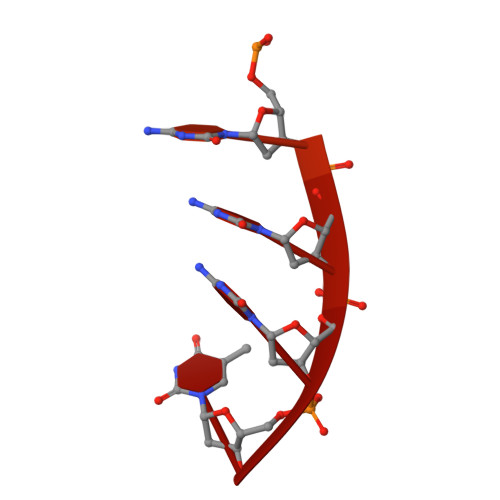
Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW Domains.
Brucet, M., Querol-Audi, J., Serra, M., Ramirez-Espain, X., Bertlik, K., Ruiz, L., Lloberas, J., Macias, M.J., Fita, I., Celada, A.(2007) J Biol Chem 282: 14547-14557
- PubMed: 17355961
- DOI: https://doi.org/10.1074/jbc.M700236200
- Primary Citation of Related Structures:
2O4G, 2O4I - PubMed Abstract:
TREX1 is the most abundant mammalian 3' --> 5' DNA exonuclease. It has been described to form part of the SET complex and is responsible for the Aicardi-Goutières syndrome in humans. Here we show that the exonuclease activity is correlated to the binding preferences toward certain DNA sequences. In particular, we have found three motifs that are selected, GAG, ACA, and CTGC. To elucidate how the discrimination occurs, we determined the crystal structures of two murine TREX1 complexes, with a nucleotide product of the exonuclease reaction, and with a single-stranded DNA substrate. Using confocal microscopy, we observed TREX1 both in nuclear and cytoplasmic subcellular compartments. Remarkably, the presence of TREX1 in the nucleus requires the loss of a C-terminal segment, which we named leucine-rich repeat 3. Furthermore, we detected the presence of a conserved proline-rich region on the surface of TREX1. This observation points to interactions with proline-binding domains. The potential interacting motif "PPPVPRPP" does not contain aromatic residues and thus resembles other sequences that select SH3 and/or Group 2 WW domains. By means of nuclear magnetic resonance titration experiments, we show that, indeed, a polyproline peptide derived from the murine TREX1 sequence interacted with the WW2 domain of the elongation transcription factor CA150. Co-immunoprecipitation studies confirmed this interaction with the full-length TREX1 protein, thereby suggesting that TREX1 participates in more functional complexes than previously thought.
Organizational Affiliation:
Macrophage Biology Group, Institute for Research in Biomedicine, University of Barcelona, Barcelona, Spain.