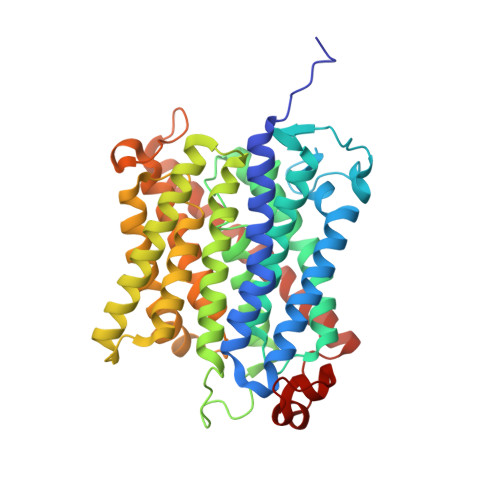
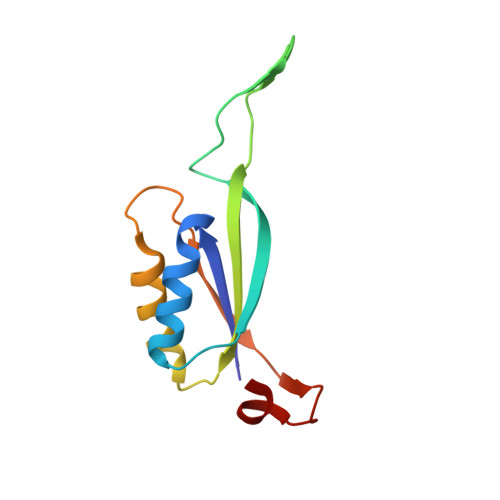
The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel
Conroy, M.J., Durand, A., Lupo, D., Li, X.-D., Bullough, P.A., Winkler, F.K., Merrick, M.(2007) Proc Natl Acad Sci U S A 104: 1213-1218
- PubMed: 17220269
- DOI: https://doi.org/10.1073/pnas.0610348104
- Primary Citation of Related Structures:
2NUU - PubMed Abstract:
Amt proteins are ubiquitous channels for the conduction of ammonia in archaea, eubacteria, fungi, and plants. In Escherichia coli, previous studies have indicated that binding of the PII signal transduction protein GlnK to the ammonia channel AmtB regulates the channel thereby controlling ammonium influx in response to the intracellular nitrogen status. Here, we describe the crystal structure of the complex between AmtB and GlnK at a resolution of 2.5 A. This structure of PII in a complex with one of its targets reveals physiologically relevant conformations of both AmtB and GlnK. GlnK interacts with AmtB almost exclusively via a long surface loop containing Y51 (T-loop), the tip of which inserts deeply into the cytoplasmic pore exit, blocking ammonia conduction. Y51 of GlnK is also buried in the pore exit, explaining why uridylylation of this residue prevents complex formation.
Organizational Affiliation:
Department of Molecular Biology and Biotechnology, University of Sheffield, Firth Court, Western Bank, Sheffield S10 2TN, United Kingdom.