C-terminal helix folding upon pyrimidine-rich hairpin binding to PTB RRM1. Implications for PTB function in Encephalomyocarditis virus IRES activity.
Maris, C., Jayne, S.F., Damberger, F.F., Ravindranathan, S., Allain, F.H.-T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polypyrimidine tract-binding protein 1 | 123 | Homo sapiens | Mutation(s): 0 Gene Names: PTB, PTBP1 | 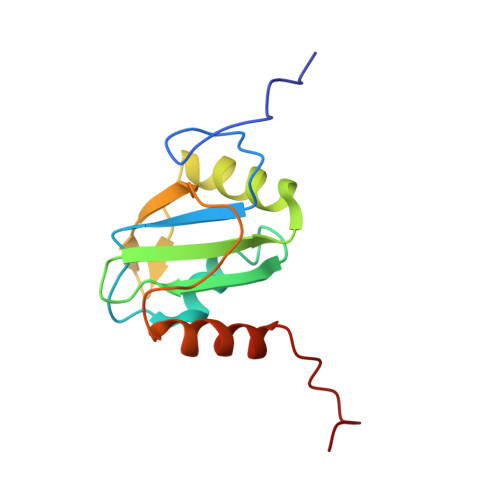 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P26599 (Homo sapiens) Explore P26599 Go to UniProtKB: P26599 | |||||
PHAROS: P26599 GTEx: ENSG00000011304 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P26599 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-R(*GP*GP*GP*AP*CP*CP*UP*GP*GP*UP*CP*UP*UP*UP*CP*CP*AP*GP*GP*UP*CP*CP*C)-3') | 23 | synthetic construct | 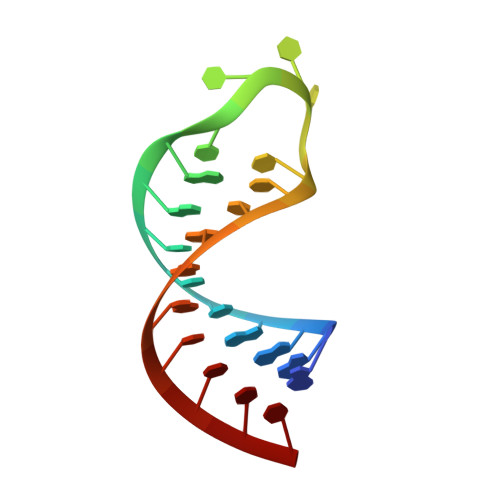 | ||
Sequence AnnotationsExpand | |||||
| |||||