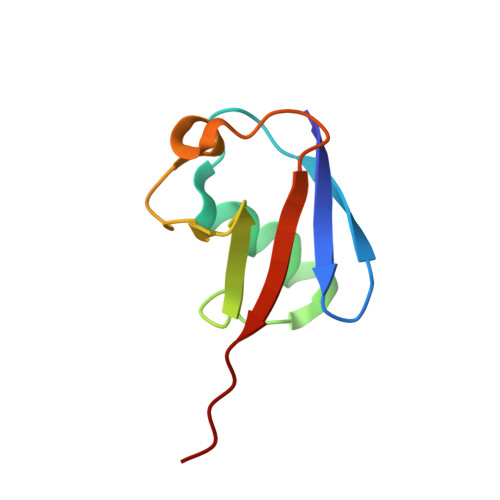
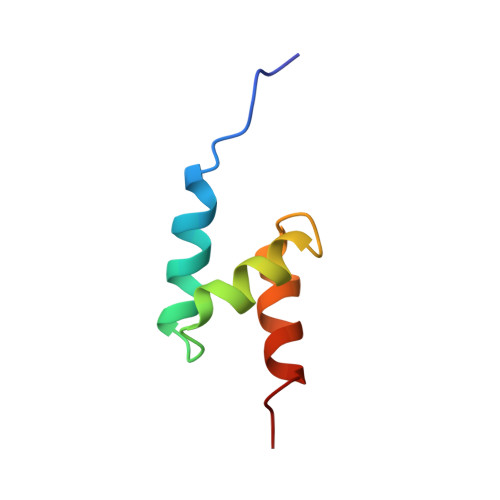
Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
Walinda, E., Morimoto, D., Sugase, K., Konuma, T., Tochio, H., Shirakawa, M.(2014) J Biol Chem 289: 13890-13902
- PubMed: 24692539
- DOI: https://doi.org/10.1074/jbc.M114.555441
- Primary Citation of Related Structures:
2MGW, 2MJ5 - PubMed Abstract:
NBR1 (neighbor of BRCA1 gene 1) is a protein commonly found in ubiquitin-positive inclusions in neurodegenerative diseases. Due to its high architectural similarity to the well studied autophagy receptor protein p62/SQSTM1, NBR1 has been thought to analogously bind to ubiquitin-marked autophagic substrates via its C-terminal ubiquitin-associated (UBA) domain and deliver them to autophagosomes for degradation. Unexpectedly, we find that NBR1 differs from p62 in its UBA structure and accordingly in its interaction with ubiquitin. Structural differences are observed on helix α-3, which is tilted farther from helix α-2 and extended by approximately one turn in NBR1. This results not only in inhibition of a p62-type self-dimerization of NBR1 UBA but also in a significantly higher affinity for monoubiquitin as compared with p62 UBA. Importantly, the NBR1 UBA-ubiquitin complex structure shows that the negative charge of the side chain in front of the conserved MGF motif in the UBA plays an integral role in the recognition of ubiquitin. In addition, NMR and isothermal titration calorimetry experiments show that NBR1 UBA binds to each monomeric unit of polyubiquitin with similar affinity and by the same surface used for binding to monoubiquitin. This indicates that NBR1 lacks polyubiquitin linkage-type specificity, in good agreement with the nonspecific linkages observed in intracellular ubiquitin-positive inclusions. Consequently, our results demonstrate that the structural differences between NBR1 UBA and p62 UBA result in a much higher affinity of NBR1 for ubiquitin, which in turn suggests that NBR1 may form intracellular inclusions with ubiquitylated autophagic substrates more efficiently than p62.
Organizational Affiliation:
From the Department of Molecular Engineering, Graduate School of Engineering, Kyoto University, Kyoto 615-8510, Japan and.