Sea Anemone Peptide with Uncommon beta-Hairpin Structure Inhibits Acid-sensing Ion Channel 3 (ASIC3) and Reveals Analgesic Activity.
Osmakov, D.I., Kozlov, S.A., Andreev, Y.A., Koshelev, S.G., Sanamyan, N.P., Sanamyan, K.E., Dyachenko, I.A., Bondarenko, D.A., Murashev, A.N., Mineev, K.S., Arseniev, A.S., Grishin, E.V.(2013) J Biol Chem 288: 23116-23127
- PubMed: 23801332
- DOI: https://doi.org/10.1074/jbc.M113.485516
- Primary Citation of Related Structures:
2LZO - PubMed Abstract:
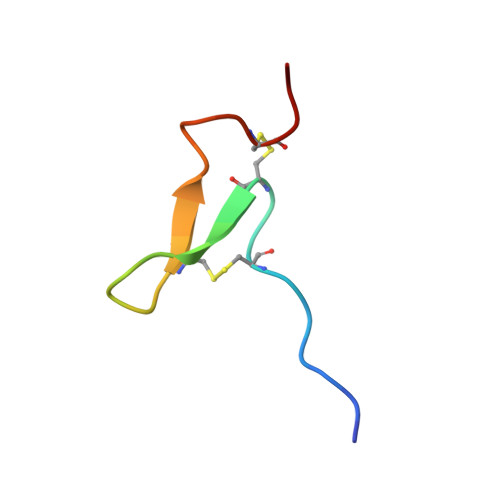
Three novel peptides were isolated from the venom of the sea anemone Urticina grebelnyi. All of them are 29 amino acid peptides cross-linked by two disulfide bridges, with a primary structure similar to other sea anemone peptides belonging to structural group 9a. The structure of the gene encoding the shared precursor protein of the identified peptides was determined. One peptide, π-AnmTX Ugr 9a-1 (short name Ugr 9-1), produced a reversible inhibition effect on both the transient and the sustained current of human ASIC3 channels expressed in Xenopus laevis oocytes. It completely blocked the transient component (IC50 10 ± 0.6 μM) and partially (48 ± 2%) inhibited the amplitude of the sustained component (IC50 1.44 ± 0.19 μM). Using in vivo tests in mice, Ugr 9-1 significantly reversed inflammatory and acid-induced pain. The other two novel peptides, AnmTX Ugr 9a-2 (Ugr 9-2) and AnmTX Ugr 9a-3 (Ugr 9-3), did not inhibit the ASIC3 current. NMR spectroscopy revealed that Ugr 9-1 has an uncommon spatial structure, stabilized by two S-S bridges, with three classical β-turns and twisted β-hairpin without interstrand disulfide bonds. This is a novel peptide spatial structure that we propose to name boundless β-hairpin.
Organizational Affiliation:
Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, ul. Miklukho-Maklaya 16/10, 117997 Moscow.