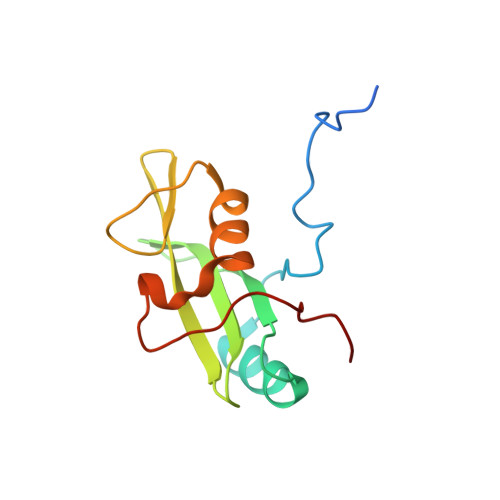
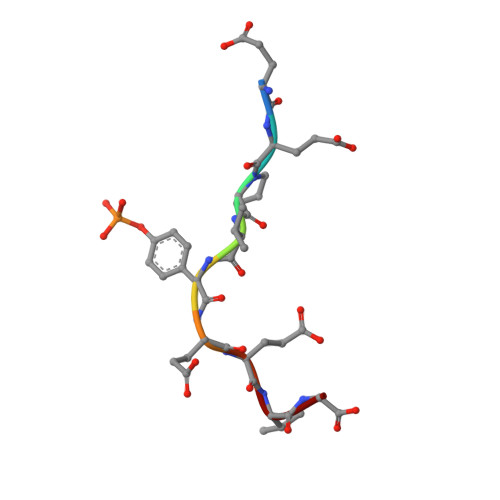
Identification and structural basis for a novel interaction between Vav2 and Arap3.
Wu, B., Wang, F., Zhang, J., Zhang, Z., Qin, L., Peng, J., Li, F., Liu, J., Lu, G., Gong, Q., Yao, X., Wu, J., Shi, Y.(2012) J Struct Biol 180: 84-95
- PubMed: 22750419
- DOI: https://doi.org/10.1016/j.jsb.2012.06.011
- Primary Citation of Related Structures:
2LNW, 2LNX - PubMed Abstract:
Vav2 is a ubiquitous guanine nucleotide exchange factor (GEF) for the small GTPase Rac1. It regulates processes including cell migration, neuronal development and phagocytosis through interactions with different proteins. In this study, Arap3, a dual GTPase-activating protein (GAP) for RhoA and Arf6, was first identified to be a novel interaction partner for Vav2 both in vitro and in vivo. ITC and NMR chemical shift perturbation experiments demonstrated that Vav2 SH2 domain was able to interact directly with phosphorylated Y1403 and Y1408 within the C-terminal region of Arap3 with high affinities, with the dissociation constants (Kd) of ≈ 0.27 and ≈ 1.40 μM, respectively. In addition, using different phosphotyrosine peptides, the pY +3 specificity of Vav2 SH2 domain was discovered. The solution structures of Vav2 SH2 domain in free and in complex with the phosphotyrosine peptide pY1408 were therefore determined to understand the structural basis of this recognition specificity. Structural analysis revealed that the presence of a Phe residue in the BG loop (BG6) leads to the formation of a shallow hydrophobic pY +3 pocket on the surface of Vav2 SH2 domain, which determines the pY +3 specificity of Vav2 SH2 domain.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, PR China.