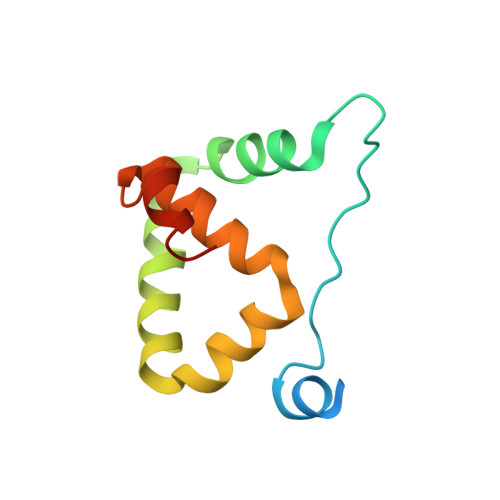
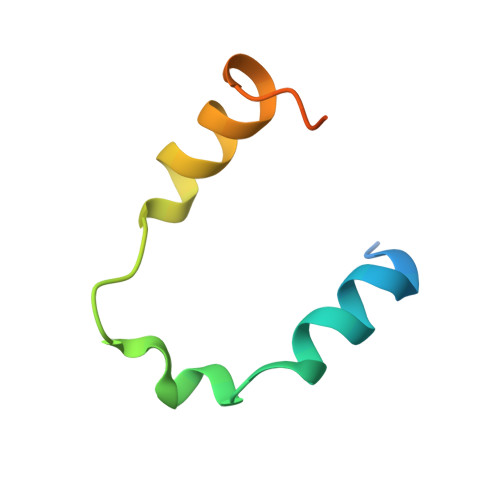
Locked Tether Formation by Cooperative Folding of Rna14p Monkeytail and Rna15p Hinge Domains in the Yeast CF IA Complex.
Moreno-Morcillo, M., Minvielle-Sebastia, L., Fribourg, S., Mackereth, C.D.(2011) Structure 19: 534-545
- PubMed: 21481776
- DOI: https://doi.org/10.1016/j.str.2011.02.003
- Primary Citation of Related Structures:
2L9B - PubMed Abstract:
The removal of the 3' region of pre-mRNA followed by polyadenylation is a key step in mRNA maturation. In the yeast Saccharomyces cerevisiae, one component of the processing machinery is the cleavage/polyadenylation factor IA (CF IA) complex, composed of four proteins (Clp1p, Pcf11p, Rna14p, Rna15p) that recognize RNA sequences adjacent to the cleavage site and recruit additional processing factors. To gain insight into the molecular architecture of CF IA we solved the solution structure of the heterodimer composed of the interacting regions between Rna14p and Rna15p. The C-terminal monkeytail domain from Rna14p and the hinge region from Rna15p display a coupled binding and folding mechanism, where both peptides are initially disordered. Mutants with destabilized monkeytail-hinge interactions prevent association of Rna15p within CF IA. Conservation of interdomain residues reveals that the structural tethering is preserved in the homologous mammalian cleavage stimulation factor (CstF)-77 and CstF-64 proteins of the CstF complex.
Organizational Affiliation:
Institut Européen de Chimie et Biologie and INSERM U869, 33607 Pessac Cedex, France.