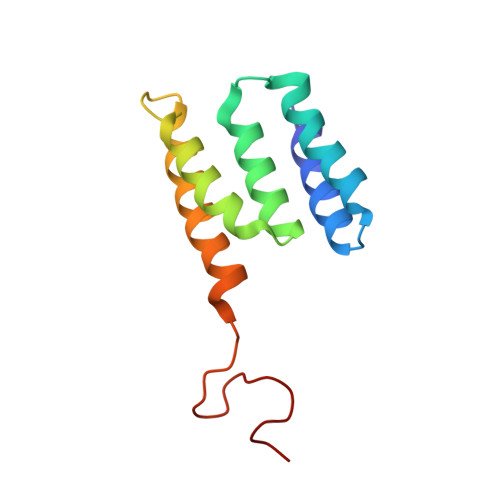
Structure of minimal tetratricopeptide repeat domain protein Tah1 reveals mechanism of its interaction with Pih1 and Hsp90.
Jimenez, B., Ugwu, F., Zhao, R., Orti, L., Makhnevych, T., Pineda-Lucena, A., Houry, W.A.(2012) J Biol Chem 287: 5698-5709
- PubMed: 22179618
- DOI: https://doi.org/10.1074/jbc.M111.287458
- Primary Citation of Related Structures:
2L6J - PubMed Abstract:
Tah1 and Pih1 are novel Hsp90 interactors. Tah1 acts as a cofactor of Hsp90 to stabilize Pih1. In yeast, Hsp90, Tah1, and Pih1 were found to form a complex that is required for ribosomal RNA processing through their effect on box C/D small nucleolar ribonucleoprotein assembly. Tah1 is a minimal tetratricopeptide repeat protein of 111 amino acid residues that binds to the C terminus of the Hsp90 molecular chaperone, whereas Pih1 consists of 344 residues of unknown fold. The NMR structure of Tah1 has been solved, and this structure shows the presence of two tetratricopeptide repeat motifs followed by a C helix and an unstructured region. The binding of Tah1 to Hsp90 is mediated by the EEVD C-terminal residues of Hsp90, which bind to a positively charged channel formed by Tah1. Five highly conserved residues, which form a two-carboxylate clamp that tightly interacts with the ultimate Asp-0 residue of the bound peptide, are also present in Tah1. Tah1 was found to bind to the C terminus of Pih1 through the C helix and the unstructured region. The C terminus of Pih1 destabilizes the protein in vitro and in vivo, whereas the binding of Tah1 to Pih1 allows for the formation of a stable complex. Based on our data, a model for an Hsp90-Tah1-Pih1 ternary complex is proposed.
Organizational Affiliation:
Structural Biochemistry Laboratory, Medicinal Chemistry Department, Centro de Investigación Príncipe Felipe, E-46012 Valencia, Spain.