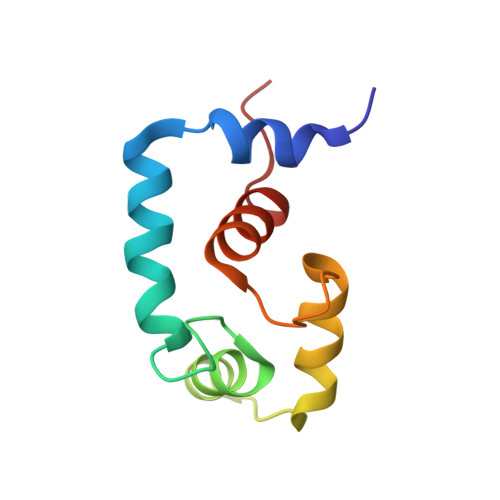
A structural and functional perspective into the mechanism of Ca(2+)-sensitizers that target the cardiac troponin complex.
Robertson, I.M., Sun, Y.B., Li, M.X., Sykes, B.D.(2010) J Mol Cell Cardiol 49: 1031-1041
- PubMed: 20801130
- DOI: https://doi.org/10.1016/j.yjmcc.2010.08.019
- Primary Citation of Related Structures:
2L1R - PubMed Abstract:
The Ca(2+) dependent interaction between troponin I (cTnI) and troponin C (cTnC) triggers contraction in heart muscle. Heart failure is characterized by a decrease in cardiac output, and compounds that increase the sensitivity of cardiac muscle to Ca(2+) have therapeutic potential. The Ca(2+)-sensitizer, levosimendan, targets cTnC; however, detailed understanding of its mechanism has been obscured by its instability. In order to understand how this class of positive inotropes function, we investigated the mode of action of two fluorine containing novel analogs of levosimendan; 2',4'-difluoro(1,1'-biphenyl)-4-yloxy acetic acid (dfbp-o) and 2',4'-difluoro(1,1'-biphenyl)-4-yl acetic acid (dfbp). The affinities of dfbp and dfbp-o for the regulatory domain of cTnC were measured in the absence and presence of cTnI by NMR spectroscopy, and dfbp-o was found to bind more strongly than dfbp. Dfbp-o also increased the affinity of cTnI for cTnC. Dfbp-o increased the Ca(2+)-sensitivity of demembranated cardiac trabeculae in a manner similar to levosimendan. The high resolution NMR solution structure of the cTnC-cTnI-dfbp-o ternary complex showed that dfbp-o bound at the hydrophobic interface formed by cTnC and cTnI making critical interactions with residues such as Arg147 of cTnI. In the absence of cTnI, docking localized dfbp-o to the same position in the hydrophobic groove of cTnC. The structural and functional data reveal that the levosimendan class of Ca(2+)-sensitizers work by binding to the regulatory domain of cTnC and stabilizing the pivotal cTnC-cTnI regulatory unit via a network of hydrophobic and electrostatic interactions, in contrast to the destabilizing effects of antagonists such as W7 at the same interface.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada.