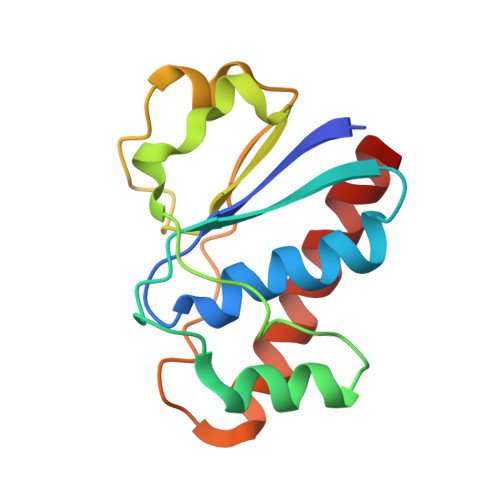
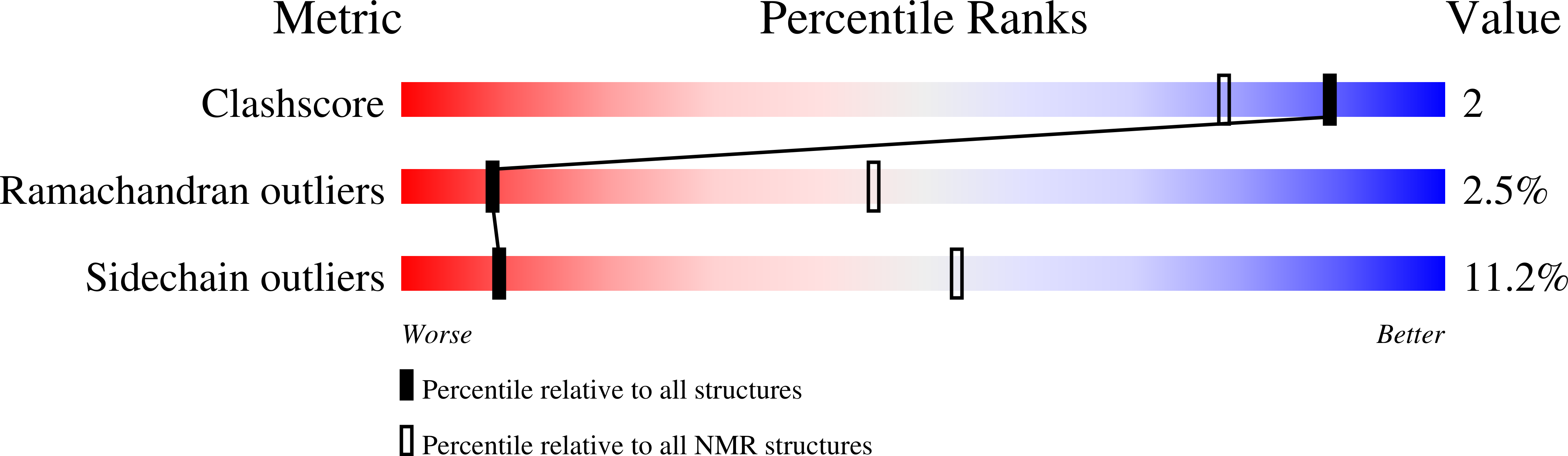(1)H, (13)C and (15)N resonance assignments of the arsenate reductase from Synechocystis sp. strain PCC 6803
Yu, C., Xia, B., Jin, C.(2011) Biomol NMR Assign 5: 85-87
- PubMed: 20960080
- DOI: https://doi.org/10.1007/s12104-010-9273-2
- Primary Citation of Related Structures:
2L17, 2L18, 2L19 - PubMed Abstract:
Arsenate reductases (ArsC) are a group of enzymes that play essential roles in biological arsenic detoxification pathways by catalyzing the intracellular reduction of arsenate to arsenite, which is subsequently extruded from the cells by specific transport systems. The ArsC protein from cyanobacterium Synechocystis sp. strain PCC 6803 (SynArsC) is related to the thioredoxin-dependent ArsC family, but uses the glutathione/glutaredoxin system for arsenate reduction. Therefore, it is classified to a novel thioredoxin/glutaredoxin hybrid arsenate reductase family. Herein we report the chemical shift assignments of (1)H, (13)C and (15)N atoms for the reduced form of SynArsC, which provides a starting point for further structural analysis and elucidation of its enzymatic mechanism.
Organizational Affiliation:
Beijing Nuclear Magnetic Resonance Center, Peking University, China.