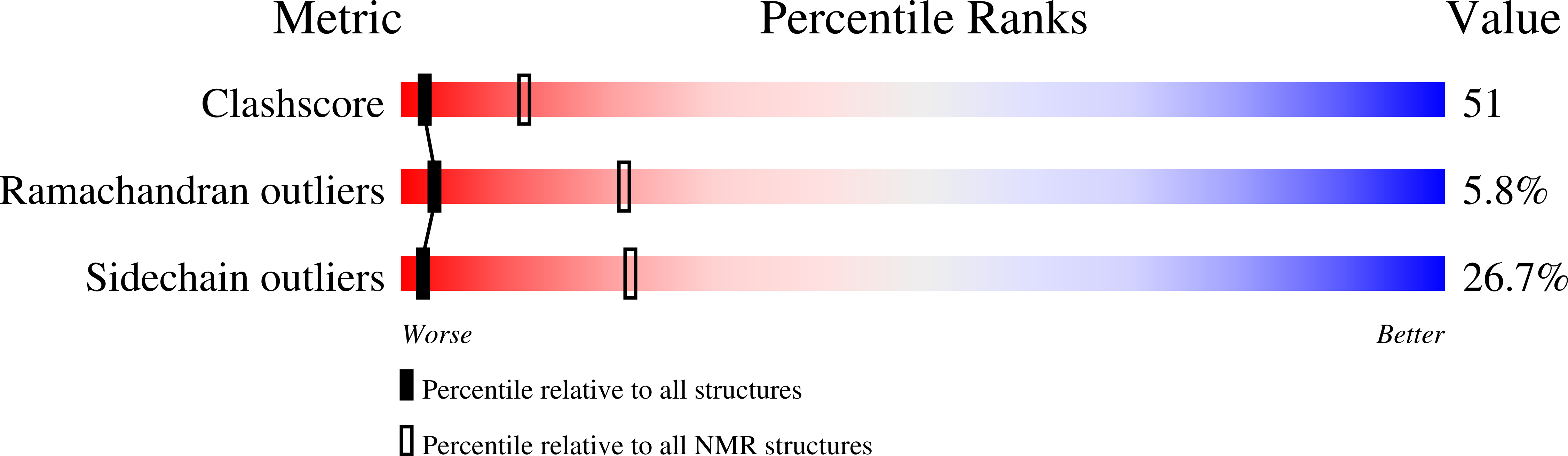Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity.
Chong, P.A., Lin, H., Wrana, J.L., Forman-Kay, J.D.(2010) Proc Natl Acad Sci U S A 107: 18404-18409
- PubMed: 20937913
- DOI: https://doi.org/10.1073/pnas.1003023107
- Primary Citation of Related Structures:
2KXQ - PubMed Abstract:
Smad ubiquitination regulatory factor 2 (Smurf2) is an E3 ubiquitin ligase that participates in degradation of TGF-β receptors and other targets. Smurf2 WW domains recognize PPXY (PY) motifs on ubiquitin ligase target proteins or on adapters, such as Smad7, that bind to E3 target proteins. We previously demonstrated that the isolated WW3 domain of Smurf2, but not the WW2 domain, can directly bind to a Smad7 PY motif. We show here that the WW2 augments this interaction by binding to the WW3 and making auxiliary contacts with the PY motif and a novel E/D-S/T-P motif, which is N-terminal to all Smad PY motifs. The WW2 likely enhances the selectivity of Smurf2 for the Smad proteins. NMR titrations confirm that Smad1 and Smad2 are bound by Smurf2 with the same coupled WW domain arrangement used to bind Smad7. The analogous WW domains in the short isoform of Smurf1 recognize the Smad7 PY peptide using the same coupled mechanism. However, a longer Smurf1 isoform, which has an additional 26 residues in the inter-WW domain linker, is only partially able to use the coupled WW domain binding mechanism. The longer linker results in a decrease in affinity for the Smad7 peptide. Interdomain coupling of WW domains enhances selectivity and enables the tuning of interactions by isoform switching.
Organizational Affiliation:
Program in Molecular Structure and Function, Hospital for Sick Children, 555 University Avenue, Toronto, ON, Canada M5G 1X8.