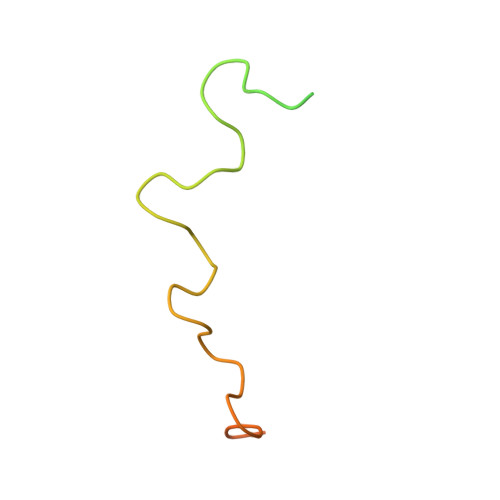
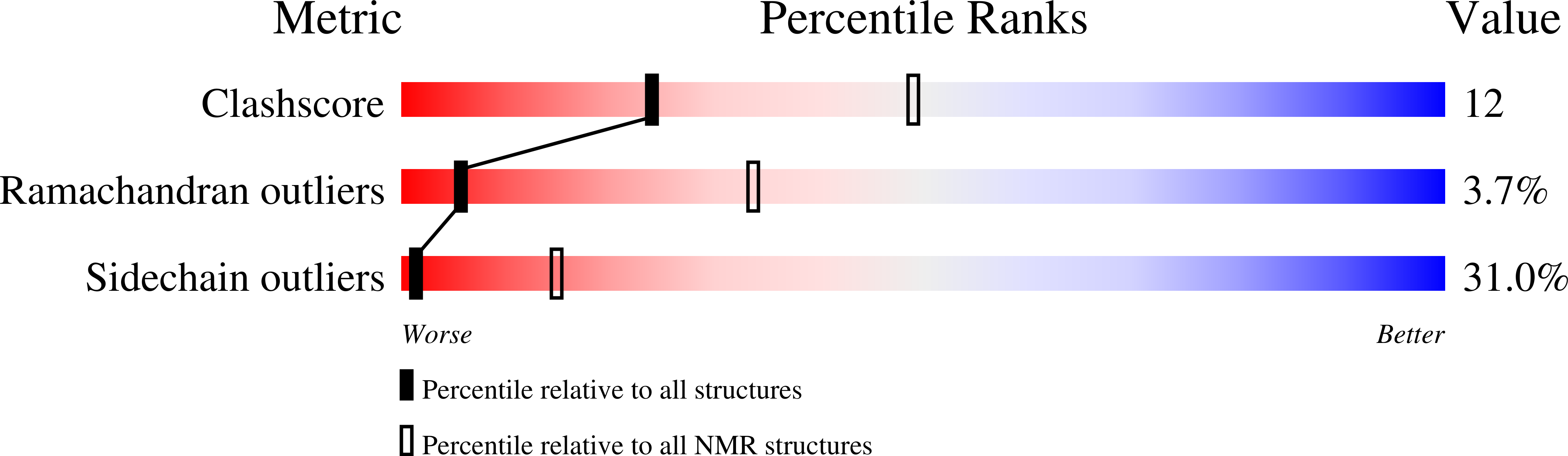Structure of the flexible amino-terminal domain of prion protein bound to a sulfated glycan.
Taubner, L.M., Bienkiewicz, E.A., Copie, V., Caughey, B.(2010) J Mol Biol 395: 475-490
- PubMed: 19913031
- DOI: https://doi.org/10.1016/j.jmb.2009.10.075
- Primary Citation of Related Structures:
2KKG - PubMed Abstract:
The intrinsically disordered amino-proximal domain of hamster prion protein (PrP) contains four copies of a highly conserved octapeptide sequence, PHGGGWGQ, that is flanked by two polycationic residue clusters. This N-terminal domain mediates the binding of sulfated glycans, which can profoundly influence the conversion of PrP to pathological forms and the progression of prion disease. To investigate the structural consequences of sulfated glycan binding, we performed multidimensional heteronuclear ((1)H, (13)C, (15)N) NMR (nuclear magnetic resonance), circular dichroism (CD), and fluorescence studies on hamster PrP residues 23-106 (PrP 23-106) and fragments thereof when bound to pentosan polysulfate (PPS). While the majority of PrP 23-106 remain disordered upon PPS binding, the octarepeat region adopts a repeating loop-turn structure that we have determined by NMR. The beta-like turns within the repeats are corroborated by CD data demonstrating that these turns are also present, although less pronounced, without PPS. Binding to PPS exposes a hydrophobic surface composed of aligned tryptophan side chains, the spacing and orientation of which are consistent with a self-association or ligand binding site. The unique tryptophan motif was probed by intrinsic tryptophan fluorescence, which displayed enhanced fluorescence of PrP 23-106 when bound to PPS, consistent with the alignment of tryptophan side chains. Chemical-shift mapping identified binding sites on PrP 23-106 for PPS, which include the octarepeat histidine and an N-terminal basic cluster previously linked to sulfated glycan binding. These data may in part explain how sulfated glycans modulate PrP conformational conversions and oligomerizations.
Organizational Affiliation:
Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA. taubnerl@niaid.nih.gov