Structure, function, and chemical synthesis of Vaejovis mexicanus peptide 24: a novel potent blocker of Kv1.3 potassium channels of human T lymphocytes.
Gurrola, G.B., Hernandez-Lopez, R.A., Rodriguez de la Vega, R.C., Varga, Z., Batista, C.V., Salas-Castillo, S.P., Panyi, G., del Rio-Portilla, F., Possani, L.D.(2012) Biochemistry 51: 4049-4061
- PubMed: 22540187
- DOI: https://doi.org/10.1021/bi300060n
- Primary Citation of Related Structures:
2K9O - PubMed Abstract:
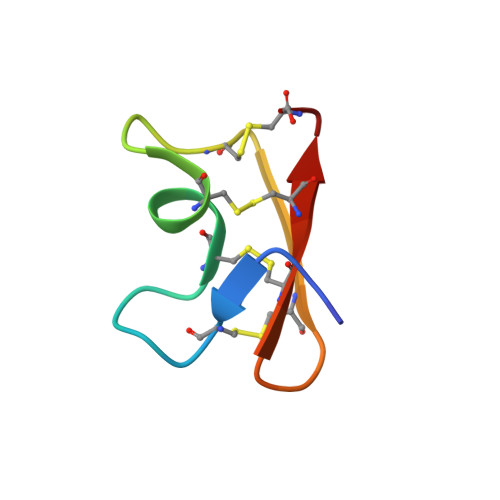
Animal venoms are rich sources of ligands for studying ion channels and other pharmacological targets. Proteomic analyses of the soluble venom from the Mexican scorpion Vaejovis mexicanus smithi showed that it contains more than 200 different components. Among them, a 36-residue peptide with a molecular mass of 3864 Da (named Vm24) was shown to be a potent blocker of Kv1.3 of human lymphocytes (K(d) ∼ 3 pM). The three-dimensional solution structure of Vm24 was determined by nuclear magnetic resonance, showing the peptide folds into a distorted cystine-stabilized α/β motif consisting of a single-turn α-helix and a three-stranded antiparallel β-sheet, stabilized by four disulfide bridges. The disulfide pairs are formed between Cys6 and Cys26, Cys12 and Cys31, Cys16 and Cys33, and Cys21 and Cys36. Sequence analyses identified Vm24 as the first example of a new subfamily of α-type K(+) channel blockers (systematic number α-KTx 23.1). Comparison with other Kv1.3 blockers isolated from scorpions suggests a number of structural features that could explain the remarkable affinity and specificity of Vm24 toward Kv1.3 channels of lymphocytes.
Organizational Affiliation:
Departamento de Medicina Molecular y Bioprocesos, Instituto de Biotecnología, Universidad Nacional Autónoma de México, Av. Universidad, 2001 Cuernavaca, Mor. 62210, Mexico.