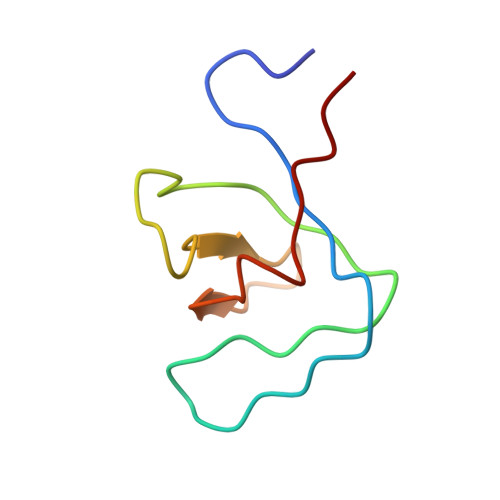
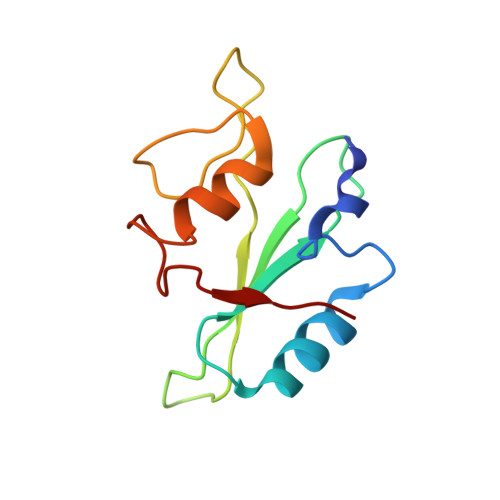
Proline isomerization preorganizes the Itk SH2 domain for binding to the Itk SH3 domain.
Severin, A., Joseph, R.E., Boyken, S., Fulton, D.B., Andreotti, A.H.(2009) J Mol Biol 387: 726-743
- PubMed: 19361414
- DOI: https://doi.org/10.1016/j.jmb.2009.02.012
- Primary Citation of Related Structures:
2K79, 2K7A - PubMed Abstract:
We report here the NMR-derived structure of the binary complex formed by the interleukin-2 tyrosine kinase (Itk) Src homology 3 (SH3) and Src homology 2 (SH2) domains. The interaction is independent of both a phosphotyrosine motif and a proline-rich sequence, the classical targets of the SH2 and SH3 domains, respectively. The Itk SH3/SH2 structure reveals the molecular details of this nonclassical interaction and provides a clear picture for how the previously described prolyl cis/trans isomerization present in the Itk SH2 domain mediates SH3 binding. The higher-affinity cis SH2 conformer is preorganized to form a hydrophobic interface with the SH3 domain. The structure also provides insight into how autophosphorylation in the Itk SH3 domain might increase the affinity of the intermolecular SH3/SH2 interaction. Finally, we can compare this Itk complex with other examples of SH3 and SH2 domains engaging their ligands in a nonclassical manner. These small binding domains exhibit a surprising level of diversity in their binding repertoires.
Organizational Affiliation:
Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University, Ames, IA 50010, USA.