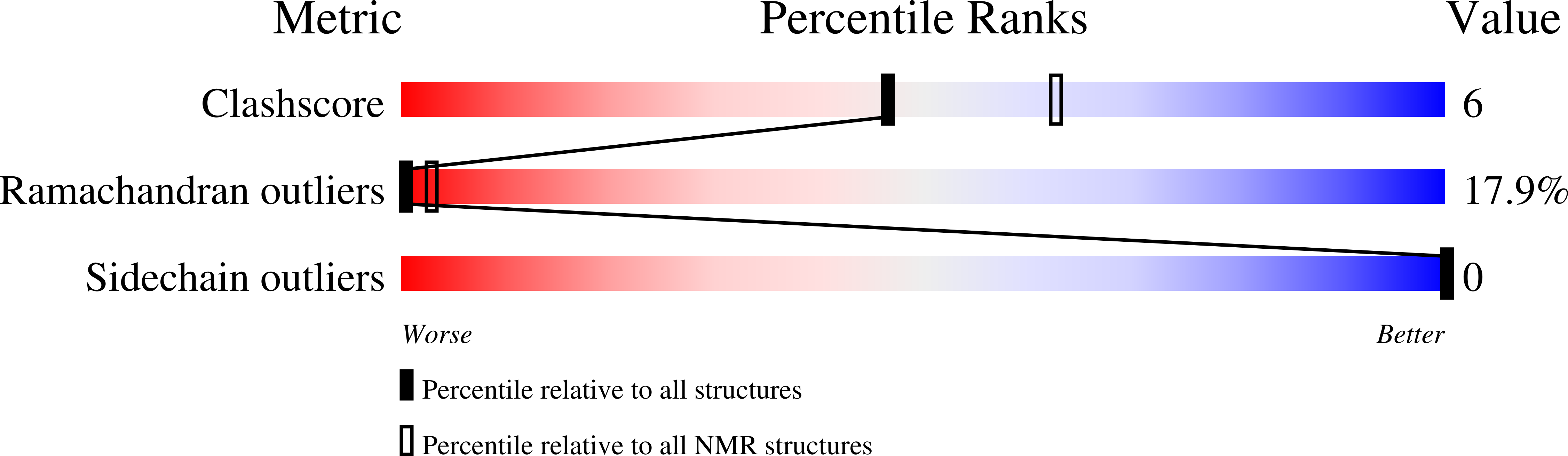Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the gp41 Fusion Peptide.
Munch, J., Standker, L., Adermann, K., Schulz, A., Schindler, M., Chinnadurai, R., Pohlmann, S., Chaipan, C., Biet, T., Peters, T., Meyer, B., Wilhelm, D., Lu, H., Jing, W., Jiang, S., Forssmann, W.G., Kirchhoff, F.(2007) Cell 129: 263-275
- PubMed: 17448989
- DOI: https://doi.org/10.1016/j.cell.2007.02.042
- Primary Citation of Related Structures:
2JNR - PubMed Abstract:
A variety of molecules in human blood have been implicated in the inhibition of HIV-1. However, it remained elusive which circulating natural compounds are most effective in controlling viral replication in vivo. To identify natural HIV-1 inhibitors we screened a comprehensive peptide library generated from human hemofiltrate. The most potent fraction contained a 20-residue peptide, designated VIRUS-INHIBITORY PEPTIDE (VIRIP), corresponding to the C-proximal region of alpha1-antitrypsin, the most abundant circulating serine protease inhibitor. We found that VIRIP inhibits a wide variety of HIV-1 strains including those resistant to current antiretroviral drugs. Further analysis demonstrated that VIRIP blocks HIV-1 entry by interacting with the gp41 fusion peptide and showed that a few amino acid changes increase its antiretroviral potency by two orders of magnitude. Thus, as a highly specific natural inhibitor of the HIV-1 gp41 fusion peptide, VIRIP may lead to the development of another class of antiretroviral drugs.
Organizational Affiliation:
Institute of Virology, University of Ulm, 89081 Ulm, Germany.