Structural Characterization of the Bradyzoite Surface Antigen (Bsr4) from Toxoplasma Gondii, a Unique Addition to the Surface Antigen Glycoprotein 1-Related Superfamily.
Crawford, J., Grujic, O., Bruic, E., Czjzek, M., Grigg, M.E., Boulanger, M.J.(2009) J Biol Chem 284: 9192
- PubMed: 19155215
- DOI: https://doi.org/10.1074/jbc.M808714200
- Primary Citation of Related Structures:
2JKS - PubMed Abstract:
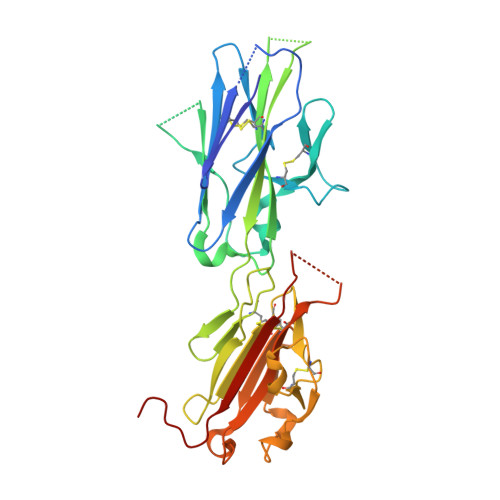
Toxoplasma gondii is an obligate intracellular protozoan parasite that infects nearly one-third of the human population. The success of T. gondii is based on its complex life cycle; a lytic tachyzoite form disseminates infection, whereas an encysted bradyzoite form establishes a latent, chronic infection. Persistence and transmissibility is central to the survival of the parasite and is, in part, mediated by a family of antigenically distinct surface antigen glycoprotein (SAG)-related sequences (SRS) adhesins that play a dual role in host cell attachment and host immune evasion. More than 160 members of the SRS family have been identified with only the tachyzoite-expressed SAG1 structurally characterized. Here we report the first structural description of the bradyzoite adhesin BSR4 using x-ray crystallography and small angle x-ray scattering. The 1.90-A crystal structure of BSR4 reveals an architecture comprised of tandem beta sandwich domains organized in a head to tail fashion with the N-terminal domain responsible for dimer formation. A restructured topology in BSR4 results in a ligand-binding site that is significantly reorganized in both structure and chemistry relative to SAG1, consistent with BSR4 binding a distinct physiological ligand. The small angle x-ray scattering solution structure of BSR4 highlights a potentially important structural role for the interdomain polymorphic linker that imparts significant flexibility that may promote structural adaptation during ligand binding. This study reveals an unexpected level of structural diversity within the SRS superfamily and provides important insight into the role of these virulence factors.
Organizational Affiliation:
Department of Biochemistry & Microbiology, University of Victoria, Victoria, British Columbia V8W 3P6, Canada.