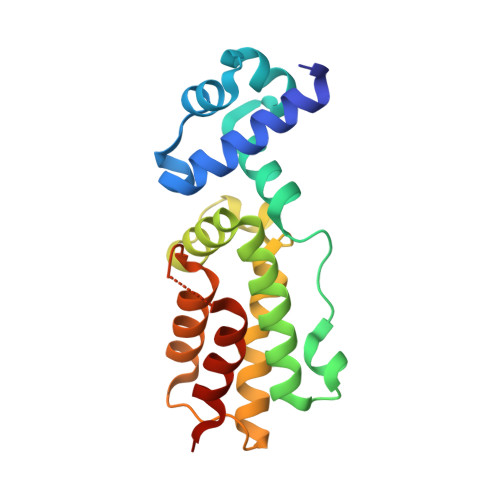
[Contraction of the disordered loop located within C-terminal domain of the transcriptional regulator HlyIIR causes its structural rearrangement].
Kovalevskii, O.V., Antson, A.A., Solonin, A.S.(2009) Mol Biol (Mosk) 43: 126-135
- PubMed: 19334535
- Primary Citation of Related Structures:
2JJ7 - PubMed Abstract:
HlyIIR is a negative transcriptional regulator of the hemolysin II gene from Bacillus cereus . A disordered region (amino acid residues 170-185) localized within the C-terminal domain near the dimerization interface was found in the recently determined HlyIIR X-ray structure. To clarify the effect of this region on HlyIIR properties and potential improvement of the diffraction quality of its crystals, we constructed a HlyIIR mutant with a single alanine residue substituting for the overall disordered region. According to biochemical analysis, the mutant protein still formed a dimer but lost its DNA-binding activity. Its crystals displayed better diffraction quality as compared with the native protein. The mutant structure was determined by X-ray analysis with a resolution of 2.1 Å. However, the mutant protein formed an alternative dimer differing from the wild-type dimer, as its subunits were rotated by 160. The conformation of individual subunits also partially changed. As this considerable remodeling in the mutant protein structure resulted from the conformational changes in the segment Pro161-Ser169, we concluded that this segment was important for maintaining the native HlyIIR structure.