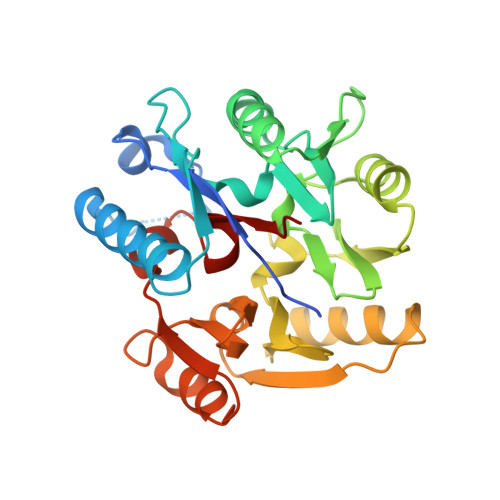
Disruption of methylarginine metabolism impairs vascular homeostasis.
Leiper, J., Nandi, M., Torondel, B., Murray-Rust, J., Malaki, M., O'Hara, B., Rossiter, S., Anthony, S., Madhani, M., Selwood, D., Smith, C., Wojciak-Stothard, B., Rudiger, A., Stidwill, R., McDonald, N.Q., Vallance, P.(2007) Nat Med 13: 198-203
- PubMed: 17273169
- DOI: https://doi.org/10.1038/nm1543
- Primary Citation of Related Structures:
2JAI, 2JAJ - PubMed Abstract:
Asymmetric dimethylarginine (ADMA) and monomethyl arginine (L-NMMA) are endogenously produced amino acids that inhibit all three isoforms of nitric oxide synthase (NOS). ADMA accumulates in various disease states, including renal failure, diabetes and pulmonary hypertension, and its concentration in plasma is strongly predictive of premature cardiovascular disease and death. Both L-NMMA and ADMA are eliminated largely through active metabolism by dimethylarginine dimethylaminohydrolase (DDAH) and thus DDAH dysfunction may be a crucial unifying feature of increased cardiovascular risk. However, despite considerable interest in this pathway and in the role of ADMA as a cardiovascular risk factor, there is little evidence to support a causal role of ADMA in pathophysiology. Here we reveal the structure of human DDAH-1 and probe the function of DDAH-1 both by deleting the DDAH1 gene in mice and by using DDAH-specific inhibitors which, as we demonstrate by crystallography, bind to the active site of human DDAH-1. We show that loss of DDAH-1 activity leads to accumulation of ADMA and reduction in NO signaling. This in turn causes vascular pathophysiology, including endothelial dysfunction, increased systemic vascular resistance and elevated systemic and pulmonary blood pressure. Our results also suggest that DDAH inhibition could be harnessed therapeutically to reduce the vascular collapse associated with sepsis.
Organizational Affiliation:
Centre for Clinical Pharmacology, Division of Medicine, University College London, London WC1E 6JJ, UK.