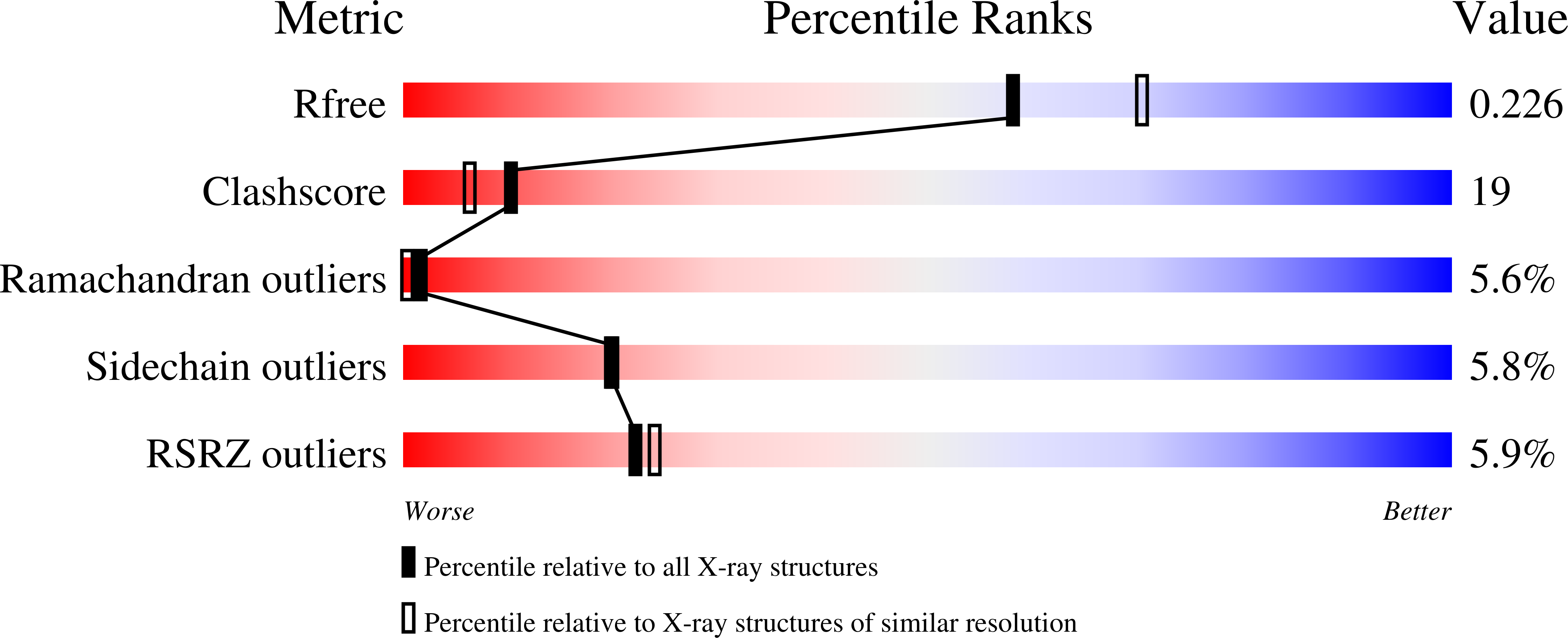
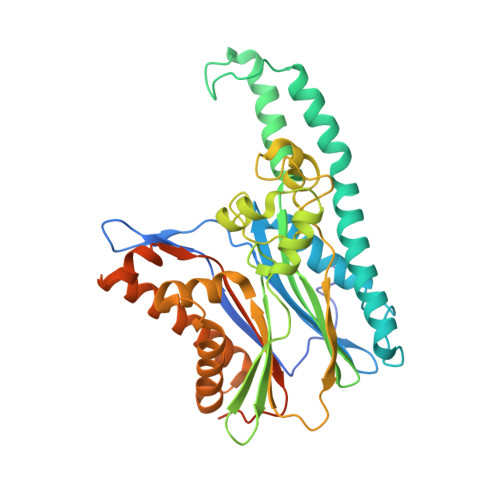
Tak1-Binding Protein 1 is a Pseudophosphatase.
Conner, S.H., Kular, G., Peggie, M., Shepherd, S., Schuttelkopf, A.W., Cohen, P., Van Aalten, D.M.F.(2006) Biochem J 399: 427
- PubMed: 16879102
- DOI: https://doi.org/10.1042/BJ20061077
- Primary Citation of Related Structures:
2J4O - PubMed Abstract:
TAB1 [TAK1 (transforming growth factor-beta-activated kinase 1)-binding protein 1] is one of the regulatory subunits of TAK1, a protein kinase that lies at the head of three pro-inflammatory kinase cascades. In the current study we report the crystal structure of the N-terminal domain of TAB1. Surprisingly, TAB1 possesses a fold closely related to that of the PPM (Mg2+- or Mn2+-dependent protein phosphatase) family as demonstrated by the close structural similarity with protein phosphatase 2C alpha. However, we were unable to detect any phosphatase activity for TAB1 using a phosphopeptide or p-nitrophenyl phosphate as substrate. Although the overall protein phosphatase 2C alpha fold is conserved in TAB1, detailed structural analyses and mutagenesis studies show that several key residues required for dual metal-binding and catalysis are not present in TAB1, although binding of a single metal is supported by soaking experiments with manganese and isothermal titration calorimetry. Thus, it appears that TAB1 is a 'pseudophosphatase', possibly binding to and regulating accessibility of phosphorylated residues on substrates downstream of TAK1 or on the TAK1 complex itself.
Organizational Affiliation:
MRC Protein Phosphorylation Unit, School of Life Sciences, University of Dundee, Dundee DD1 5EH, Scotland, UK.