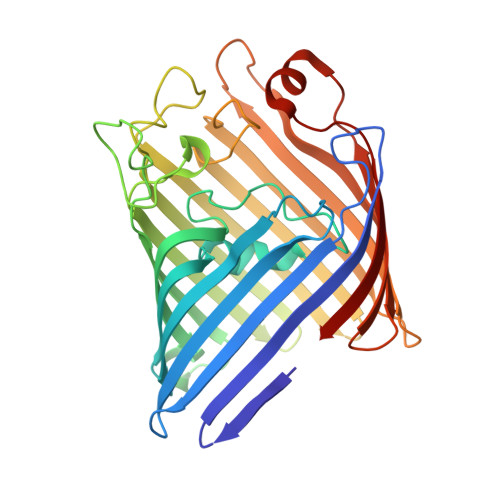
Crystal Structure of Osmoporin Ompc from E. Coli at 2.0 A.
Basle, A., Rummel, G., Storici, P., Rosenbusch, J.P., Schirmer, T.(2006) J Mol Biol 362: 933
- PubMed: 16949612
- DOI: https://doi.org/10.1016/j.jmb.2006.08.002
- Primary Citation of Related Structures:
2J1N - PubMed Abstract:
Porins form transmembrane pores in the outer membrane of Gram-negative bacteria with matrix porin OmpF and osmoporin OmpC from Escherichia coli being differentially expressed depending on environmental conditions. The three-dimensional structure of OmpC has been determined to 2.0 A resolution by X-ray crystallography. As expected from the high sequence similarity, OmpC adopts the OmpF-like 16-stranded hollow beta-barrel fold with three beta-barrels associated to form a tight trimer. Unlike in OmpF, the extracellular loops form a continuous wall at the perimeter of the vestibule common to the three pores, due to a 14-residues insertion in loop L4. The pore constriction and the periplasmic outlet are very similar to OmpF with 74% of the pore lining residues being conserved. Overall, only few ionizable residues are exchanged at the pore lining. The OmpC structure suggests that not pore size, but electrostatic pore potential and particular atomic details of the pore linings are the critical parameters that physiologically distinguish OmpC from OmpF.
Organizational Affiliation:
Division of Structural Biology, University of Basel, Klingelbergstrasse 70, CH-4056 Basel, Switzerland.