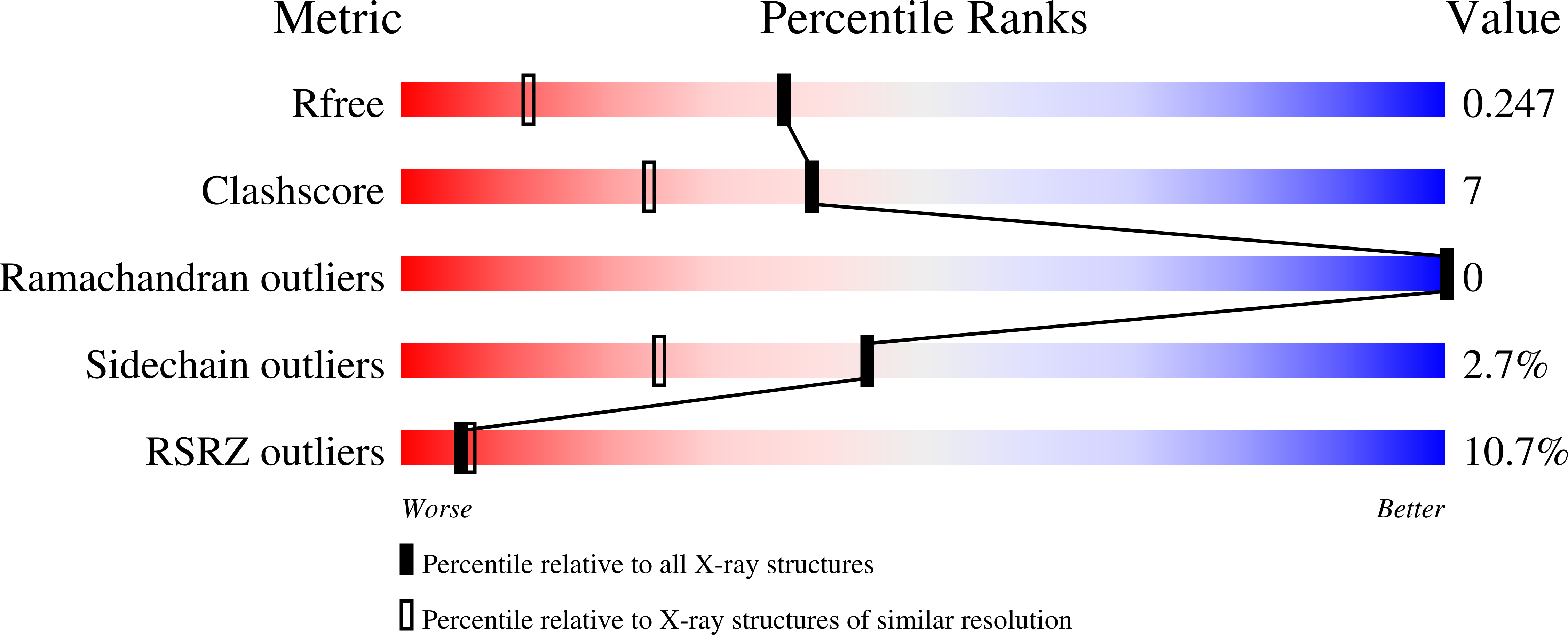
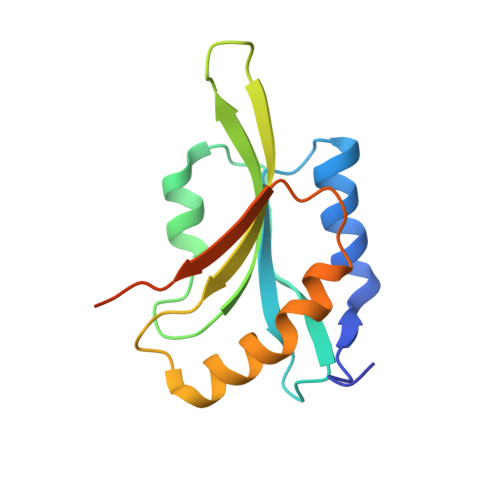
Mechanism of Actin Filament Turnover by Severing and Nucleation at Different Concentrations of ADF/Cofilin.
Andrianantoandro, E., Pollard, T.D.(2006) Mol Cell 24: 13-23
- PubMed: 17018289
- DOI: https://doi.org/10.1016/j.molcel.2006.08.006
- Primary Citation of Related Structures:
2I2Q - PubMed Abstract:
ADF/cofilins are key regulators of actin dynamics during cellular motility, yet their precise role and mechanism of action are shrouded in ambiguity. Direct observation of actin filaments by evanescent wave microscopy showed that cofilins from fission yeast and human do not increase the rate that pointed ends of actin filaments shorten beyond the rate for ADP-actin subunits, but both cofilins inhibit elongation and subunit dissociation at barbed ends. Direct observation also showed that cofilins from fission yeast, Acanthamoeba, and human sever actin filaments optimally at low-cofilin binding densities well below their K(d)s, but not at high binding densities. High concentrations of cofilin nucleate actin assembly. Thus, the action of cofilins in cells will depend on the local concentration of active cofilins: low concentrations favor severing, whereas high concentrations favor nucleation. These results establish a clear paradigm for actin turnover by cofilin in cells.
Organizational Affiliation:
Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520, USA.