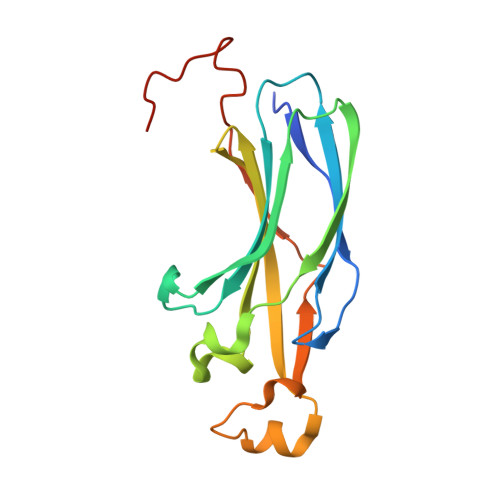
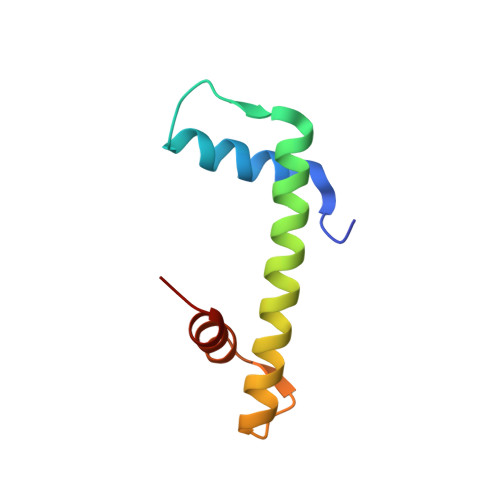
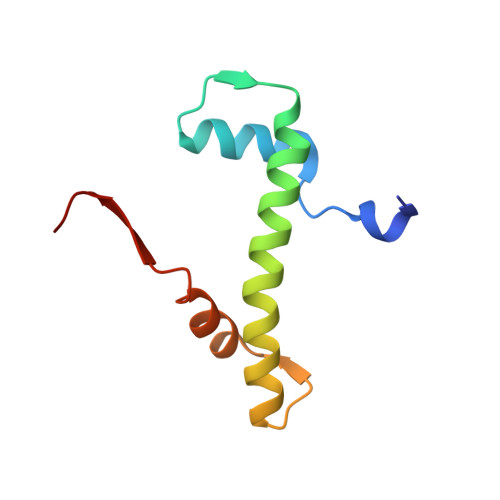
Structural basis for the histone chaperone activity of asf1.
English, C.M., Adkins, M.W., Carson, J.J., Churchill, M.E., Tyler, J.K.(2006) Cell 127: 495-508
- PubMed: 17081973
- DOI: https://doi.org/10.1016/j.cell.2006.08.047
- Primary Citation of Related Structures:
2HUE - PubMed Abstract:
Anti-silencing function 1 (Asf1) is a highly conserved chaperone of histones H3/H4 that assembles or disassembles chromatin during transcription, replication, and repair. The structure of the globular domain of Asf1 bound to H3/H4 determined by X-ray crystallography to a resolution of 1.7 Angstroms shows how Asf1 binds the H3/H4 heterodimer, enveloping the C terminus of histone H3 and physically blocking formation of the H3/H4 heterotetramer. Unexpectedly, the C terminus of histone H4 that forms a mini-beta sheet with histone H2A in the nucleosome undergoes a major conformational change upon binding to Asf1 and adds a beta strand to the Asf1 beta sheet sandwich. Interactions with both H3 and H4 were required for Asf1 histone chaperone function in vivo and in vitro. The Asf1-H3/H4 structure suggests a "strand-capture" mechanism whereby the H4 tail acts as a lever to facilitate chromatin disassembly/assembly that may be used ubiquitously by histone chaperones.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, School of Medicine, University of Colorado, Aurora, CO 80045, USA.