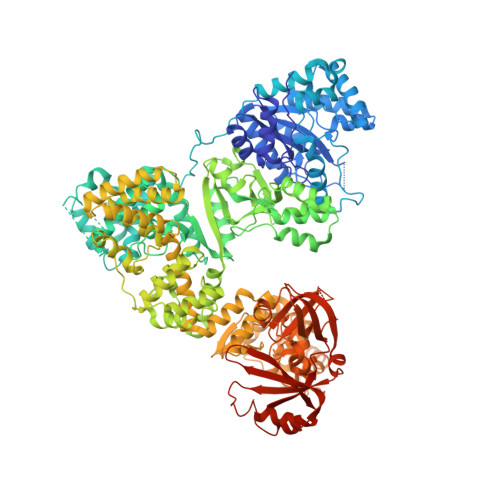
The Structure of T. aquaticus DNA Polymerase III Is Distinct from Eukaryotic Replicative DNA Polymerases.
Bailey, S., Wing, R.A., Steitz, T.A.(2006) Cell 126: 893-904
- PubMed: 16959569
- DOI: https://doi.org/10.1016/j.cell.2006.07.027
- Primary Citation of Related Structures:
2HPI, 2HPM - PubMed Abstract:
The crystal structure of Thermus aquaticus DNA polymerase III alpha subunit reveals that the structure of the catalytic domain of the eubacterial replicative polymerase is unrelated to that of the eukaryotic replicative polymerase but rather belongs to the Polbeta-like nucleotidyltransferase superfamily. A model of the polymerase complexed with both DNA and beta-sliding clamp interacting with a reoriented binding domain and internal beta binding site was constructed that is consistent with existing biochemical data. Within the crystal, two C-terminal domains are interacting through a surface that is larger than many dimer interfaces. Since replicative polymerases of eubacteria and eukaryotes/archaea are not homologous, the nature of the replicative polymerase in the last common ancestor is unknown. Although other possibilities have been proposed, the plausibility of a ribozyme DNA polymerase should be considered.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.