Gain of Function in an ERV/ALR Sulfhydryl Oxidase by Molecular Engineering of the Shuttle Disulfide.
Vitu, E., Bentzur, M., Lisowsky, T., Kaiser, C.A., Fass, D.(2006) J Mol Biol 362: 89-101
- PubMed: 16893552
- DOI: https://doi.org/10.1016/j.jmb.2006.06.070
- Primary Citation of Related Structures:
2HJ3 - PubMed Abstract:
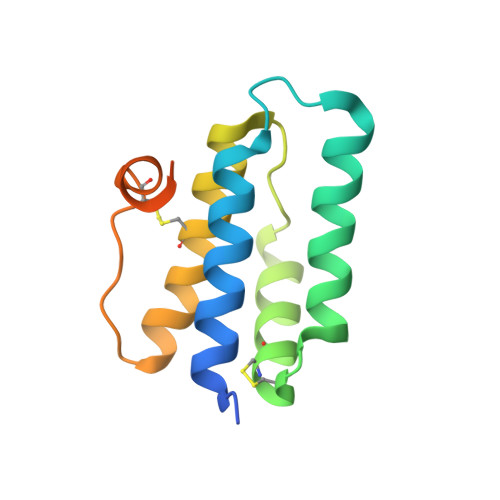
The ERV/ALR sulfhydryl oxidase domain is a versatile module adapted for catalysis of disulfide bond formation in various organelles and biological settings. Its four-helix bundle structure juxtaposes a Cys-X-X-Cys dithiol/disulfide motif with a bound flavin adenine dinucleotide (FAD) cofactor, enabling transfer of electrons from thiol substrates to non-thiol electron acceptors. ERV/ALR family members contain an additional di-cysteine motif outside the four-helix-bundle core. Although the location and context of this "shuttle" disulfide differs among family members, it is proposed to perform the same basic function of mediating electron transfer from substrate to the enzyme active site. We have determined by X-ray crystallography the structure of AtErv1, an ERV/ALR enzyme that contains a Cys-X4-Cys shuttle disulfide and oxidizes thioredoxin in vitro, and compared it to ScErv2, which has a Cys-X-Cys shuttle and does not oxidize thioredoxin at an appreciable rate. The AtErv1 shuttle disulfide is in a region of the structure that is disordered and thus apparently mobile and exposed. This feature may facilitate access of protein substrates to the shuttle disulfide. To test whether the shuttle disulfide region is modular and can confer on other enzymes oxidase activity toward new substrates, we generated chimeric enzyme variants combining shuttle disulfide and core elements from AtErv1 and ScErv2 and monitored oxidation of thioredoxin by the chimeras. We found that the AtErv1 shuttle disulfide region could indeed confer thioredoxin oxidase activity on the ScErv2 core. Remarkably, various chimeras containing the ScErv2 Cys-X-Cys shuttle disulfide were found to function efficiently as well. Since neither the ScErv2 core nor the Cys-X-Cys motif is therefore incapable of participating in oxidation of thioredoxin, we conclude that wild-type ScErv2 has evolved to repress activity on substrates of this type, perhaps in favor of a different, as yet unknown, substrate.
Organizational Affiliation:
Department of Structural Biology, Weizmann Institute of Science, Rehovot 76100, Israel.