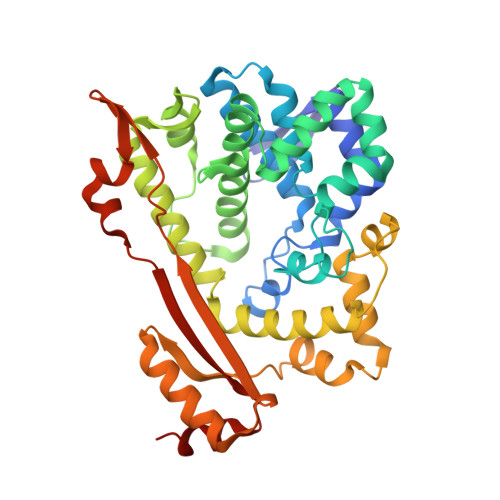
Structure of O67745_AQUAE, a hypothetical protein from Aquifex aeolicus.
Oganesyan, V., Adams, P.D., Jancarik, J., Kim, R., Kim, S.H.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 369-374
- PubMed: 17565173
- DOI: https://doi.org/10.1107/S1744309107018945
- Primary Citation of Related Structures:
2HEK - PubMed Abstract:
Using single-wavelength anomalous dispersion data obtained from a gold-derivatized crystal, the X-ray crystal structure of the protein 067745_AQUAE from the prokaryotic organism Aquifex aeolicus has been determined to a resolution of 2.0 A. Amino-acid residues 1-371 of the 44 kDa protein were identified by Pfam as an HD domain and a member of the metal-dependent phosphohydrolase superfamily (accession No. PF01966). Although three families from this large and diverse group of enzymatic proteins are represented in the PDB, the structure of 067745_AQUAE reveals a unique fold that is unlike the others and that is likely to represent a new subfamily, further organizing the families and characterizing the proteins. Data are presented that provide the first insights into the structural organization of the proteins within this clan and a distal alternative GDP-binding domain outside the metal-binding active site is proposed.
Organizational Affiliation:
Berkeley Structural Genomics Center, Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA.