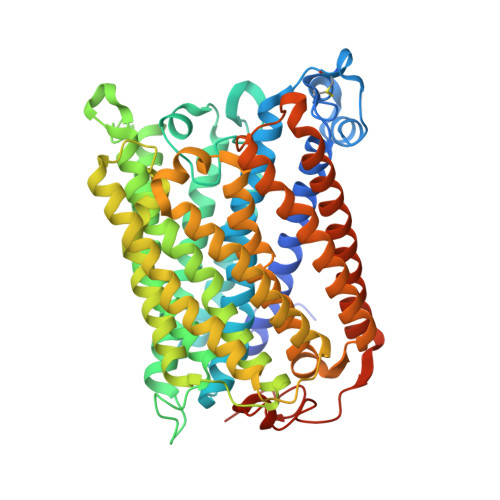
Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase.
Qin, L., Hiser, C., Mulichak, A., Garavito, R.M., Ferguson-Miller, S.(2006) Proc Natl Acad Sci U S A 103: 16117-16122
- PubMed: 17050688
- DOI: https://doi.org/10.1073/pnas.0606149103
- Primary Citation of Related Structures:
2GSM - PubMed Abstract:
Well ordered reproducible crystals of cytochrome c oxidase (CcO) from Rhodobacter sphaeroides yield a previously unreported structure at 2.0 A resolution that contains the two catalytic subunits and a number of alkyl chains of lipids and detergents. Comparison with crystal structures of other bacterial and mammalian CcOs reveals that the positions occupied by native membrane lipids and detergent substitutes are highly conserved, along with amino acid residues in their vicinity, suggesting a more prevalent and specific role of lipid in membrane protein structure than often envisioned. Well defined detergent head groups (maltose) are found associated with aromatic residues in a manner similar to phospholipid head groups, likely contributing to the success of alkyl glycoside detergents in supporting membrane protein activity and crystallizability. Other significant features of this structure include the following: finding of a previously unreported crystal contact mediated by cadmium and an engineered histidine tag; documentation of the unique His-Tyr covalent linkage close to the active site; remarkable conservation of a chain of waters in one proton pathway (D-path); and discovery of an inhibitory cadmium-binding site at the entrance to another proton path (K-path). These observations provide important insight into CcO structure and mechanism, as well as the significance of bound lipid in membrane proteins.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA.