The Crystal Structure of the Transthyretin-like Protein from Salmonella dublin, a Prokaryote 5-Hydroxyisourate Hydrolase.
Hennebry, S.C., Law, R.H., Richardson, S.J., Buckle, A.M., Whisstock, J.C.(2006) J Mol Biol 359: 1389-1399
- PubMed: 16787778
- DOI: https://doi.org/10.1016/j.jmb.2006.04.057
- Primary Citation of Related Structures:
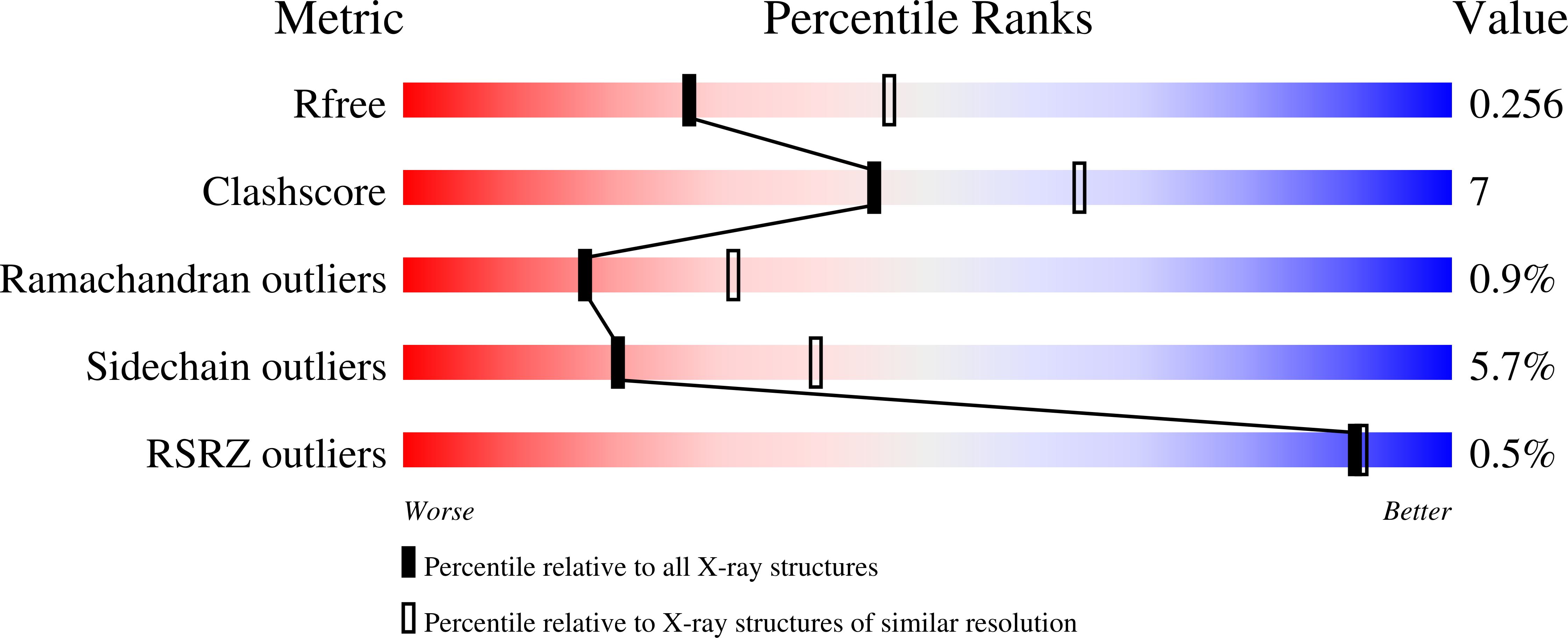
2GPZ - PubMed Abstract:
The mechanism of binding of thyroid hormones by the transport protein transthyretin (TTR) in vertebrates is structurally well characterised. However, a homologous family of transthyretin-like proteins (TLPs) present in bacteria as well as eukaryotes do not bind thyroid hormones, instead they are postulated to perform a role in the purine degradation pathway and function as 5-hydroxyisourate hydrolases. Here we describe the 2.5 Angstroms X-ray crystal structure of the TLP from the Gram-negative bacterium Salmonella dublin, and compare and contrast its structure with vertebrate TTRs. The overall architecture of the homotetramer is conserved and, despite low sequence homology with vertebrate TTRs, structural differences within the monomer are restricted to flexible loop regions. However, sequence variation at the dimer-dimer interface has profound consequences for the ligand binding site and provides a structural rationalisation for the absence of thyroid hormone binding affinity in bacterial TLPs: the deep, negatively charged thyroxine-binding pocket that characterises vertebrate TTR contrasts with a shallow and elongated, positively charged cleft in S. dublin TLP. We have demonstrated that Sdu_TLP is a 5-hydroxyisourate hydrolase. Furthermore, using site-directed mutagenesis, we have identified three conserved residues located in this cleft that are critical to the enzyme activity. Together our data reveal that the active site of Sdu_TLP corresponds to the thyroxine binding site in TTRs.
Organizational Affiliation:
The Department of Biochemistry and Molecular Biology, Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Parkville, VIC, Australia.